Create your own panel
In G2P, each dataset is focused on a disease grouping or defined category of clinical presentation that is of relevance to the clinical diagnosis of Mendelian disease. For a dataset to be publicly released it must be comprehensive, up-to-date and have a plan for future active curation. If you would like to start creating your own panel using the gene2phenotype website please contact us g2p-help@ebi.ac.uk and we will add the new panel and user accounts to our site. After the user account has been set up and the panels which can be updated are configured there are two ways of creating and populating a panel.
Firstly, you can provide us a file which contains the new gene disease entities, and we will load those entities into the database which will be available for editing through the website after loading. Please encode your gene disease entities in the same way as we do for our data dumps of existing panels available from our download page. Not all fields need to be available for an entry to be loaded into the database. The minimal information available must be gene symbol, allelic requirement, mutation consequence, disease name and gene disease association confidence value. Everything else can be entered using the gene2phenotype website.
Secondly, you can use the web interface for adding a new entry to a panel by first searching the gene2phenotype website with the gene symbol of your gene of interest. Search works with autocomplete. All Ensembl genes are preloaded with their HGNC symbol. We also store previously assigned HGNC symbols. If you cannot find your gene of interest, please contact g2p-help@ebi.ac.uk. The main entities which can be curated for a gene disease pair are: disease confidence, disease mechanism which is pair consisting of allelic requirement and mutation consequence, publications, phenotypes and organ specificity.
Once you started curating your panel you can download the annotations and use it as input for the VEP G2P plugin. If you already annotate an existing panel with PanelApp it is also possible to use the file version of your PanelApp panel and use it as input for the VEP G2P plugin.
Curator views
Create a new entry
An entry describes a gene disease association which is characterized by the allelic requirement and mutation consequence. We want to reduce the existence of duplicated data in the database as much as possible. Therefore, entries that are the same across panels are annotated together. When entering a new entry, the following information must be provided:- Gene symbol
- Disease name
- Allelic requirement
- Mutation consequence
- Target panel
- Gene disease association confidence

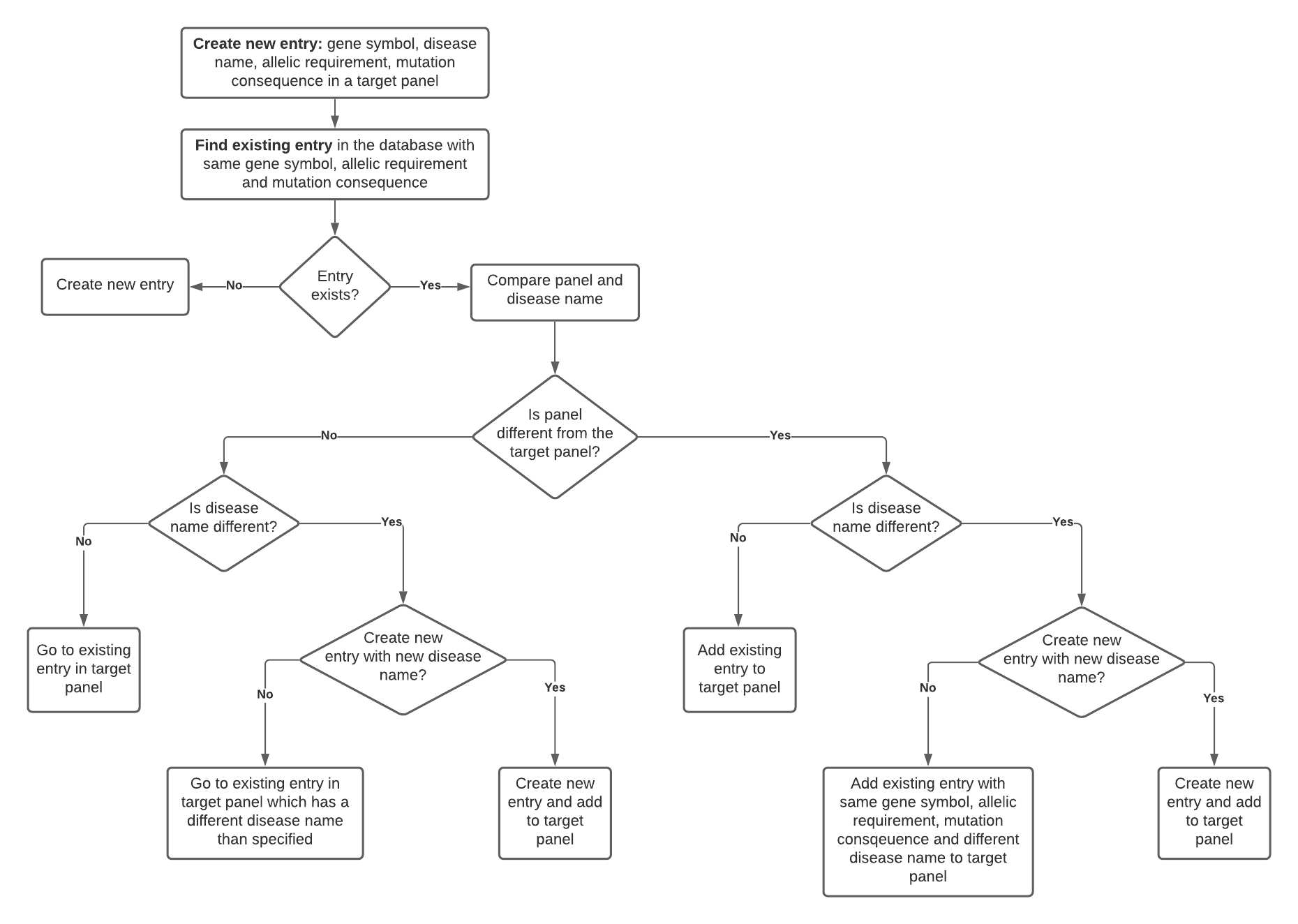
Bevor creating a new entry, we will check if there is already an entry with the same gene symbol, allelic requirement and mutation consequence in the database. Depending on the outcome of the search result the user can choose from several options which are described in a flowchart and encompass:
- The entry is already in the database, but with a different disease name.
- The user needs to decide now if the existing entry represents the new association. If it does and is already in the target panel, the user can now go to the existing entry. If it does but is not in the target panel, the user can add the existing entry to the target panel and then curate the existing entry together with curators from other panels
- The entry is already in the database, with the same disease name, but in a different panel.
- The user adds the existing entry to the target panel.
- The entry doesn’t exist, and we create a new entry in the target panel.
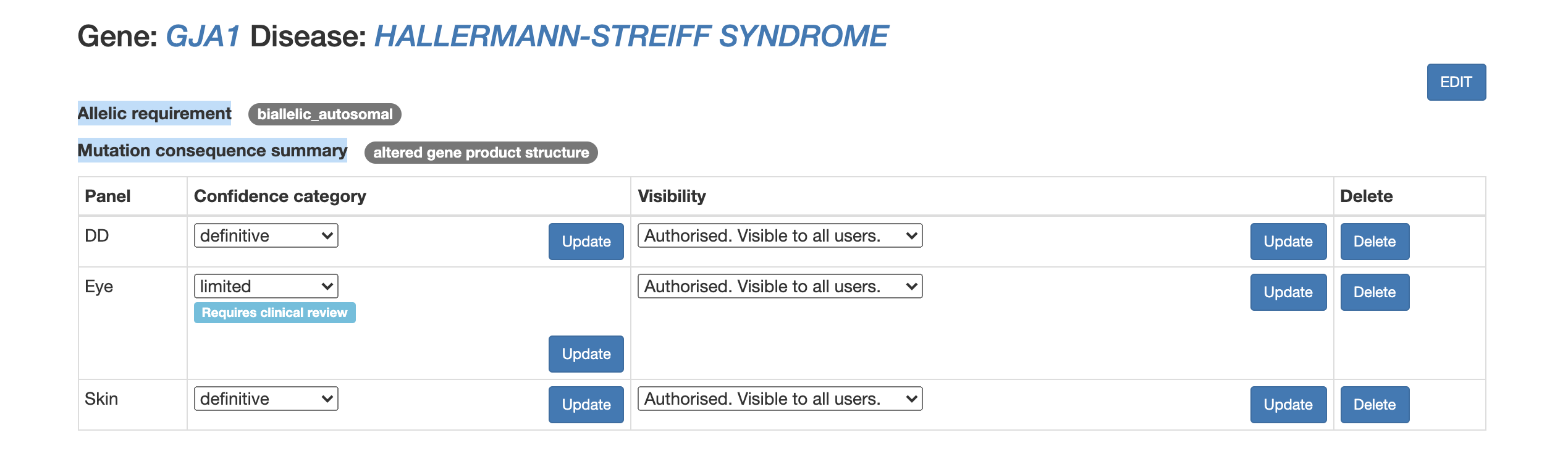
Panel specific curation
An entry can belong to several panels. The panel specific curators have permissions to add an entry to a panel and set the confidence category, visibility status and finally can delete an entry from the panel. The confidence category – confirmed, probable or possible – is assigned to indicate how likely it is that the gene is implicated in the cause of disease. The visibility setting decides if the entry can be seen as part of the panel by users that are not logged in on the website.
Publications

- We store the links to the publications (via PMID) that provide evidence for a specific gene-disease pair. A new publication is retrived by its PMID using Europe PMC's REST API.
- The title of the added publication is listed and links out to Europe PMC. A publication can be deleted and you can add comments for a publication.
Add phenotypes from HPO

- Phenotype terms are added from the Human Phenotype Ontology. The term can be selected using autocomplete.
- Phenotype terms are added to the list of phenotypes associated with the G2P entry.
- After the new phenotype term has been added to the list of phenotypes it can be deleted again or comments can be added for the phenotype.
- You can add a comment or disard and go back to the list of phenotypes.
Add organ specifity

- Selected organ specificity for a gene-disease pair.
- Edit organ specificity list.