Recently, a paper describing PDBe’s new volume slicer was published in the Journal of Structural Biology. The volume slicer is a web-based tool that allows scientists to look in detail at 3D bioimaging datasets. It is the first step in the development of an integrated browser that will allow visualisation, integration and analysis of 3D structural data on scales from molecules to cells. The development of this integrated browser is part of PDBe’s MOL2CELL project. Gerard Kleywegt tells you more.
MOL2CELL addresses several challenges in structure archiving
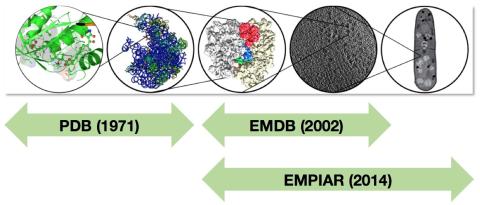
PDBe’s stated mission is to bring structure to biology. To fulfill that mission, PDBe is involved in the two major established archives in structural biology, the PDB (Protein Data Bank) and EMDB (Electron Microscopy Data Bank), and provides a plethora of services and information to the scientific community based on the data held in these and other archives. However, the science is changing rapidly and this poses challenges to the community and to archival and dissemination resources such as PDBe. These challenges include the increasing size and complexity of structures and data, the use of more heterogeneous information at a range of scales in structural studies, the coordination of archiving efforts across disciplines, the integration of structural data on scales from atoms to cells (Figure 1) and of structure with other biological and chemical data, and how to deliver appropriate structure data to non-experts (ideally in the context of their work).

Figure 1 - Structural archiving on scales from biomacromolecules (mostly in PDB), via large complexes and molecular machines (PDB and EMDB), to cellular components and beyond (EMDB and EMPIAR).
The MOL2CELL project at PDBe addresses a number of issues related to these challenges. The initial ideas sprang from a workshop in 2011 on data-management challenges in 3DEM and they were addressed further in another workshop in 2012 on the 3D cellular context of the macromolecular world. The recommendations from the latter meeting formed the basis for a grant application to the UK’s Medical Research Council (MRC). This application was successful, and is co-funded by the UK’s BBSRC research council.
In line with the recommendations of the 2012 workshop, the MOL2CELL project has three components:
(1) The establishment of a pilot archive of raw 2D data associated with EMDB entries, which can be used for validation, methods development, teaching, community challenges, etc. This has become the EMPIAR archive, which was discussed here recently.
(2) The development of tools for annotating segmentations of 3D imaging data using terms or entries from other bioinformatics resources, classifications and ontologies (such as UniProt and GO). These annotations can be used to link 3D cellular imaging data to molecular structures (determined by X-ray, NMR and EM) in PDB and EMDB, and to other biological data resources.
(3) The development of an integrated viewer for structural data on scales from atoms to cells, to make structural information on a range of scales easily accessible and thus facilitate knowledge discovery.

Figure 2 - Early mock-up of an integrated 3D structure browser, linking cellular and molecular structure data through annotated segmentations.
The new volume slicer provides easy access to complex bioimaging data
We discussed the importance of integrated access to structural data on a range of scales several years ago, and at the time we made a mock-up of what an integrated structure browser could look like (Figure 2). When the MOL2CELL project began, the first step was to develop a web-based browser for orthogonal slices through 3D imaging datasets. This work resulted in what is now called the volume slicer (Figure 3), and it is described in the recent paper in the Journal of Structural Biology. Although developed for the MOL2CELL project, feedback during user testing was enthusiastic and suggested that it would be very useful to have the volume slicer available for all the EM maps and tomograms in EMDB. (Previously, we only provided a tool to view slices of tomograms, and only perpendicular to the “Z-axis”.)

Figure 3 - The volume slicer, developed as part of the MOL2CELL project, which is now available for all maps and tomograms in EMDB. This example shows the volume slicer applied to entry EMD-2362.
Segmentations and their annotation are important to link diverse data
A critical and challenging component of the integration of cellular and molecular structure data is to find ways to link them together. Molecular structures are held in the PDB and they have rich annotation and up-to-date links to other bioinformatics resources, in the case of proteins largely provided by the SIFTS resource. Volume and image data held in EMDB and EMPIAR on the other hand rarely comes with spatial delineations and annotations of individual molecules. “Spatial delineation” means that one has indicated in 3D space the location, shape, extent and orientation of things that are “interesting” (in optical microscopy and medical imaging, these are called “regions of interest” or ROIs) - this could be cellular components, molecular machines, or individual molecules (Figure 4). One way of doing this is by generating segments (using software, manually, or using a combination of the two) - 3D volumes or “masks” that coincide with some interesting feature or object in a map or tomogram. A critical requirement is to annotate these segments with terms that signify the biological interpretation by the microscopist as to what the interesting bit represents (e.g., “inner membrane” or “polysome”). To make it possible to link such annotations to other bioinformatics resources (including the PDB), these terms should come from a well-defined and widely used vocabulary (for example the Gene Ontology, GO) or be a direct reference to an entry in a well-established resource (such as UniProt). The central panel in the mock-up viewer of Figure 2 shows examples of how such segmentations could be annotated to provide links to other archives and resources. To discuss the details of how such annotation could be derived and stored, PDBe and BioMedBridges organised a workshop in December 2015 that brought together experts in EM, tomography, segmentation and ontologies.

Figure 4. Example of a segmented EM map, in this case of a 50S ribosome with a bound protein called HslX (EMDB entry EMD-3133). The segments labeled L1, L2, etc. correspond to various ribosomal proteins. Once these segments are annotated with their corresponding UniProt identifiers, it will be easy to link them to entries in other relevant archives and databases, such as the PDB.
The MOL2CELL project is currently funded until the autumn of 2017. Watch this space for more exciting developments! For a photo of the MOL2CELL project team at PDBe, see our recent EMPIAR blog.
Reference
The paper describing the volume slicer was published as: J. Salavert-Torres, A. Iudin, I. Lagerstedt, E. Sanz-García, G.J. Kleywegt & A. Patwardhan. “Web-based volume slicer for 3D electron-microscopy data from EMDB.” Journal of Structural Biology 194, 164-170 (2016). http://dx.doi.org/10.1016/j.jsb.2016.02.012.
Acknowledgements
We are very grateful for the support from our funders, in particular MRC (grant MR/L007835, co-funded by BBSRC), BBSRC (grant BB/M018423), the Wellcome Trust (grant 104948) and EMBL-EBI, to the participants in the various workshops related to MOL2CELL, and to all the community experts who have provided input and feedback on all stages of the project.


