PDBe provides PDB-entry pages where you can easily inspect electron-density maps for all X-ray crystal structures (and it works for structures with EM maps as well), and which make it especially easy to inspect the maps for ligands (example). We take a look at the history and relevance of this functionality that is replacing the Uppsala Electron-Density Server (EDS).
Why is it useful to inspect electron density of ligands?
In the early days of the PDB, the depositors and the users of this archive were essentially the same group of people: protein crystallographers. Nowadays, however, most users of the PDB are not themselves structural biologists. In fact, a back-of-the-envelope calculation suggests that less than 10% of PDBe website users are structural biologists.
Whereas crystallographers are generally aware that not every detail of every crystal structure is accurate to 0.001 Å (which is the precision with which spatial coordinates of atoms are archived in the PDB), this is not necessarily appreciated by other scientists. Ligands, in particular, are often a very important component of a structure (and indeed often the reason for determining a structure), yet examples unfortunately abound of ligand complexes that have been published and deposited in the PDB where there is no, or at best partial or ambiguous experimental support for the conformation, location or even presence of the important ligand. For crystal structures, this experimental support can be assessed by visualising the electron-density map and checking whether it covers the entire ligand and its surroundings in a plausible manner.
Why are some maps red or green?
You are likely to encounter two kinds of maps, the so-called “2mFo-DFc” map, which should show density contours surrounding all well-determined atoms in the model, and the “mFo-DFc” map (also called difference map or difference Fourier). Negative density features in a difference map point at things that are in the model whereas the experimental data says they shouldn’t be there. These could be atoms that are either not present in the crystal or not well determined by the data, or indicators of other aspects that have not been modeled correctly. Positive features in this map, on the other hand, show aspects of the structure that are reflected in the experimental data, but not (yet) accounted for in the model. These may include missing entities such as ions or water molecules.
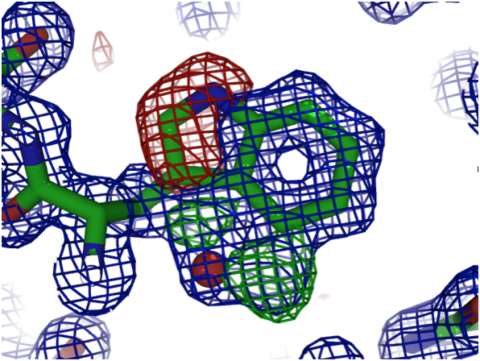
Many crystallographers use the convention of colouring the 2mFo-DFc density blue (or grey, or white), positive difference density green and negative difference density red. For instance, a sidechain that has been modeled incorrectly should ideally show “red” density around its atoms as well as “blue” and “green” density that suggest where it should be placed instead. In the figure below, you see a tryptophan sidechain with blue, green and red density, all indicating that the sidechain has not been modeled correctly. Based on these maps, you can easily figure out how it should have been modeled instead. (Bonus points if you can figure out what the red sphere inside the blue density is!)

Figure 1. Example of a crystallographic model displayed with blue 2mFo-DFc density and green (positive) and red (negative) difference density.
How can I view density maps?
The PDB has supported archiving of experimental X-ray data (in the form of structure-factor amplitudes or intensities) since its inception, but for 25 years nothing much was done with them. In the mid-1990s, Alwyn Jones and co-workers in Uppsala (Sweden) decided to expose this treasure trove of experimental data in a way that would be useful for experts and non-experts alike and would help them assess the quality of crystal structures (both overall and in the details, such as ligands). The result of this EU-funded effort was EDS (paper), a server that provides access to validation statistics and plots as well as electron-density maps for most X-ray crystal structures in the PDB (example). EDS provides a Java-based viewer for structures and maps, and several molecular graphics packages are capable of retrieving EDS data directly and showing it to their users (e.g., Chimera, PyMol and Coot). And now PDBe provides much the same functionality as EDS.
I don’t recognize the 3D viewer PDBe uses to display densities?
In 2014, PDBe began a very productive collaboration with the laboratory of Professor Jaroslav Koča at the CEITEC institute in Brno (Czech Republic) and with his colleague David Sehnal in particular. David has developed two pieces of software that enabled the development of the model-density-display functionality. First, he developed an extremely efficient coordinate server that can read a PDB structure (in mmCIF format) and serve all or selected coordinates, taking crystallographic symmetry into account. Second, he wrote a powerful yet lightweight 3D graphics component, LiteMol. Both are now used on the PDBe website to show structures and their density maps (if they are X-ray or EM structures).
What will happen to EDS now?
EDS has been running in Uppsala for almost two decades, but as the development matured and the funding ran out, it became time to move the functionality to a core bioinformatics facility, in this case PDBe at EMBL-EBI. The EDS server in Uppsala will be retired some time in 2017 (more information, especially for "power users") and we recommend all current EDS users to switch to using the PDBe functionality (and files).
In the past decade, wwPDB has convened a number of Validation Task Forces (VTFs), for X-ray, NMR and EM, which are providing recommendations on how structures determined by these methods should be validated. These recommendations are implemented in a software pipeline that produces a human-readable report as well as a file with “raw” validation statistics. PDBe is exposing more and more of the validation information available in these reports on its entry pages to further enhance their utility. Once all this is in place, most of the functionality offered by EDS will not only have been replaced, but in an improved fashion, and it is kept up-to-date on a weekly basis by PDBe. Moreover, the validation information will be integrated with all the other biological and chemical information about every archive entry.

Figure 2. The red arrows point to two different ways of viewing models and density maps on PDBe entry pages (in this case, http://pdbe.org/1cbs). If you want to see the entire model and its density, click on "3D Visualisation"; if you are interested in just a ligand and its binding site (and maps), click on its 2D structure.
Can I try out this new viewer?
The viewer is available for all PDB entries on the PDBe website. For example, by clicking on the retinoic-acid structure on the entry page for 1CBS, you can inspect the binding site and density maps for the all-trans-retinoic acid ligand (and, for comparison, visit the EDS entry for this structure). There is a quick guide that shows you how to use and interact with LiteMol. If you want to see density for things other than ligands, go to an X-ray entry’s PDBe page (example) and click on the link “3D Visualisation” in the panel on the right-hand side. By default, the density will not be shown, but as soon as you click on any atom, it will be switched on. There is also a tutorial available that teaches you more about structure validation and how you can use PDBe resources to help you assess the quality and reliability of structures in the PDB.
We would be happy to receive feedback and suggestions for improvement on the new functionality (for instance: is it intuitive to use? are there additional features you would like to see? does it work properly in your browser?). You can send feedback using the Feedback button on the PDBe entry pages. Of course, if you like the new functionality, please tell your colleagues, students, social media friends, etc. about it!

Figure 3. Part of a LiteMol display, including the set of parameters that determine the level, contouring radius, and rendering of the maps.


