Function and Biology Details
Reaction catalysed:
Cleavage of 5-His-|-Leu-6, 10-His-|-Leu-11, 14-Ala-|-Leu-15, 16-Tyr-|-Leu-17 and 23-Gly-|-Phe-24 of insulin B chain. With small molecule substrates prefers hydrophobic residue at P2' and small residue such as Ala, Gly at P1.
Biochemical function:
Biological process:
Cellular component:
- not assigned
Sequence domains:
Structure domain:
Structure analysis Details
Assembly composition:
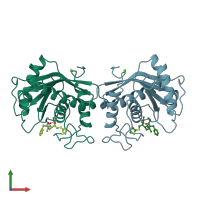
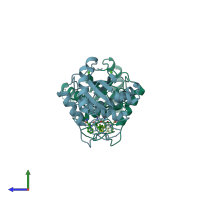
homo dimer (preferred)
Assembly name:
Snake venom metalloproteinase atrolysin-D (preferred)
PDBe Complex ID:
PDB-CPX-537876 (preferred)
Entry contents:
1 distinct polypeptide molecule
Macromolecule: