EC 1.14.99.66: [Histone-H3]-N(6),N(6)-dimethyl-L-lysine(4) FAD-dependent demethylase
Reaction catalysed:
(1a) a [histone H3]-N(6),N(6)-dimethyl-L-lysine(4) + acceptor + H(2)O = a [histone H3]-N(6)-methyl-L-lysine(4) + formaldehyde + reduced acceptor
Systematic name:
[Histone H3]-N(6),N(6)-dimethyl-L-lysine(4):acceptor oxidoreductase (demethylating)
Alternative Name(s):
- Lysine-specific histone demethylase 1
GO terms
Biochemical function:
Biological process:
Cellular component:
Sequence families
 Protein families (Pfam)
Protein families (Pfam)
 InterPro annotations
InterPro annotations
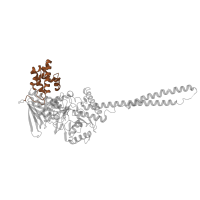
Structure domains
 CATH domains
CATH domains
1.10.287.80
Class:
Mainly Alpha
Architecture:
Orthogonal Bundle
Topology:
Helix Hairpins
Homology:
ATP synthase, gamma subunit, helix hairpin domain
Occurring in:

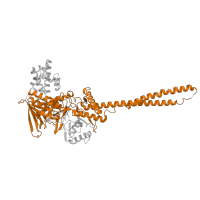
 SCOP annotations
SCOP annotations
51913
Class:
Alpha and beta proteins (a/b)
Fold:
FAD/NAD(P)-binding domain
Superfamily:
FAD/NAD(P)-binding domain
Occurring in:
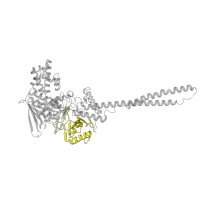
54394
Class:
Alpha and beta proteins (a+b)
Fold:
FAD-linked reductases, C-terminal domain
Superfamily:
FAD-linked reductases, C-terminal domain
Occurring in:
140222
Class:
All alpha proteins
Fold:
DNA/RNA-binding 3-helical bundle
Superfamily:
Homeodomain-like
Occurring in: