EC 2.7.7.7: DNA-directed DNA polymerase
Reaction catalysed:
Deoxynucleoside triphosphate + DNA(n) = diphosphate + DNA(n+1)
Systematic name:
Deoxynucleoside-triphosphate:DNA deoxynucleotidyltransferase (DNA-directed)
Alternative Name(s):
- DNA duplicase
- DNA nucleotidyltransferase
- DNA nucleotidyltransferase (DNA-directed)
- DNA polymerase
- DNA polymerase I
- DNA polymerase II
- DNA polymerase III
- DNA polymerase alpha
- DNA polymerase beta
- DNA polymerase gamma
- DNA replicase
- DNA-dependent DNA polymerase
- Deoxynucleate polymerase
- Deoxyribonucleate nucleotidyltransferase
- Deoxyribonucleic acid duplicase
- Deoxyribonucleic acid polymerase
- Deoxyribonucleic duplicase
- Deoxyribonucleic polymerase
- Deoxyribonucleic polymerase I
- Duplicase
- Klenow fragment
- Sequenase
- Taq DNA polymerase
- Taq Pol I
- Tca DNA polymerase
GO terms
Biochemical function:
Biological process:
Cellular component:
- not assigned
Sequence families
 Protein families (Pfam)
Protein families (Pfam)
 InterPro annotations
InterPro annotations
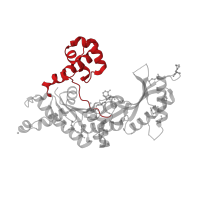
IPR036775
Domain description:
DNA polymerase, Y-family, little finger domain superfamily
Occurring in:
Occurring in:
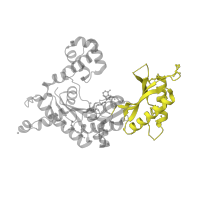
Structure domains
 CATH domains
CATH domains
3.30.70.270
Class:
Alpha Beta
Architecture:
2-Layer Sandwich
Topology:
Alpha-Beta Plaits
Homology:
Alpha-Beta Plaits
Occurring in:
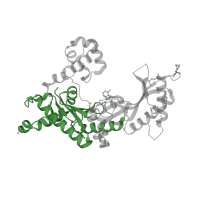
3.30.1490.100
Class:
Alpha Beta
Architecture:
2-Layer Sandwich
Topology:
Dna Ligase; domain 1
Homology:
DNA polymerase, Y-family, little finger domain
Occurring in:
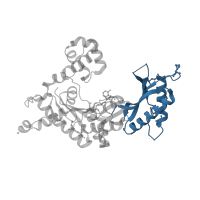
1.10.150.20
Class:
Mainly Alpha
Architecture:
Orthogonal Bundle
Topology:
DNA polymerase; domain 1
Homology:
5' to 3' exonuclease, C-terminal subdomain
Occurring in: