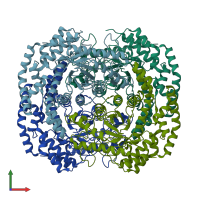
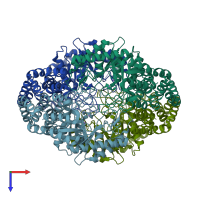
Assemblies
Assembly Name:
Xylose isomerase-like TIM barrel domain-containing protein
Multimeric state:
homo tetramer
Accessible surface area:
51406.92 Å2
Buried surface area:
26401.82 Å2
Dissociation area:
7,654.27
Å2
Dissociation energy (ΔGdiss):
67.2
kcal/mol
Dissociation entropy (TΔSdiss):
17.13
kcal/mol
Symmetry number:
4
PDBe Complex ID:
PDB-CPX-181127
Macromolecules
Chains: A, B, C, D
Length: 438 amino acids
Theoretical weight: 48.05 KDa
Source organism: Pseudomonas stutzeri
Expression system: Escherichia coli
UniProt:
Pfam: Xylose isomerase-like TIM barrel
InterPro:
Length: 438 amino acids
Theoretical weight: 48.05 KDa
Source organism: Pseudomonas stutzeri
Expression system: Escherichia coli
UniProt:
- Canonical:
 Q75WH8 (Residues: 1-430; Coverage: 100%)
Q75WH8 (Residues: 1-430; Coverage: 100%)
Pfam: Xylose isomerase-like TIM barrel
InterPro:
- L-rhamnose catabolism isomerase
- Xylose isomerase-like superfamily
- Xylose isomerase-like, TIM barrel domain