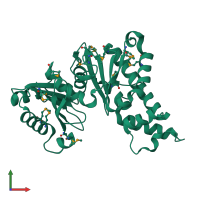
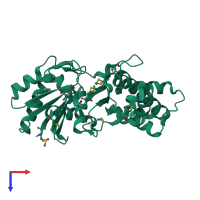
Assemblies
Assembly Name:
Hypothetical sugar kinase
Multimeric state:
homo dimer
Accessible surface area:
24860.87 Å2
Buried surface area:
3063.22 Å2
Dissociation area:
1,531.61
Å2
Dissociation energy (ΔGdiss):
15.26
kcal/mol
Dissociation entropy (TΔSdiss):
13.67
kcal/mol
Symmetry number:
2
PDBe Complex ID:
PDB-CPX-163972
Macromolecules
Chain: A
Length: 321 amino acids
Theoretical weight: 35.33 KDa
Source organism: Cytophaga hutchinsonii ATCC 33406
Expression system: Escherichia coli
CATH: Nucleotidyltransferase; domain 5
Length: 321 amino acids
Theoretical weight: 35.33 KDa
Source organism: Cytophaga hutchinsonii ATCC 33406
Expression system: Escherichia coli
CATH: Nucleotidyltransferase; domain 5