Assemblies
Assembly Name:
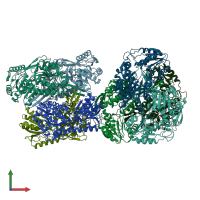
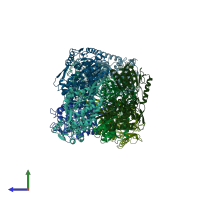
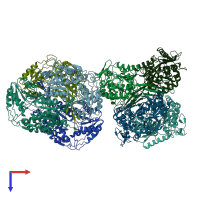
Aldehyde dehydrogenase
Multimeric state:
homo tetramer
Accessible surface area:
60341.72 Å2
Buried surface area:
15554.17 Å2
Dissociation area:
3,550.5
Å2
Dissociation energy (ΔGdiss):
42.29
kcal/mol
Dissociation entropy (TΔSdiss):
16.25
kcal/mol
Symmetry number:
4
PDBe Complex ID:
PDB-CPX-235861
Assembly Name:
Aldehyde dehydrogenase
Multimeric state:
homo tetramer
Accessible surface area:
60345.96 Å2
Buried surface area:
15411.53 Å2
Dissociation area:
3,478.44
Å2
Dissociation energy (ΔGdiss):
38.78
kcal/mol
Dissociation entropy (TΔSdiss):
16.24
kcal/mol
Symmetry number:
4
PDBe Complex ID:
PDB-CPX-235861
Macromolecules
Chains: A, B, C, D, E, F, G, H
Length: 516 amino acids
Theoretical weight: 55.05 KDa
Source organism: Pseudomonas putida S12
Expression system: Escherichia coli
Length: 516 amino acids
Theoretical weight: 55.05 KDa
Source organism: Pseudomonas putida S12
Expression system: Escherichia coli
- Homing endonuclease
- Aldehyde/histidinol dehydrogenase
- Aldehyde dehydrogenase domain
- Aldehyde dehydrogenase, N-terminal
- Aldehyde dehydrogenase, glutamic acid active site
- Aldehyde dehydrogenase, C-terminal
- Aldehyde dehydrogenase, cysteine active site
RSRZ Outlier Chain A (auth A)
Chain A (auth A)
RSRZ Outlier Chain B (auth B)
Chain B (auth B)
RSRZ Outlier Chain C (auth C)
Chain C (auth C)
RSRZ Outlier Chain D (auth D)
Chain D (auth D)
RSRZ Outlier Chain E (auth E)
Chain E (auth E)
RSRZ Outlier Chain F (auth F)
Chain F (auth F)
RSRZ Outlier Chain G (auth G)
Chain G (auth G)
RSRZ Outlier Chain H (auth H)
Chain H (auth H)
Domains
Secondary structure
Flexibility predictions
Early folding residue predictions
Sequence conservation