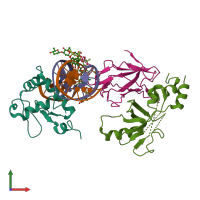
Function and Biology Details
Biochemical function:
Biological process:
Cellular component:
Sequence domains:
- Ets domain
- Acute myeloid leukemia 1 protein (AML1)/Runt
- Core-binding factor, beta subunit superfamily
- Core-binding factor, beta subunit
- Runt domain
- p53-like transcription factor, DNA-binding domain superfamily
- p53/RUNT-type transcription factor, DNA-binding domain superfamily
- Winged helix DNA-binding domain superfamily
2 more domains
Structure analysis Details
Assembly composition:
hetero pentamer (preferred)
PDBe Complex ID:
PDB-CPX-120775 (preferred)
Entry contents:
3 distinct polypeptide molecules
2 distinct DNA molecules
2 distinct DNA molecules
Macromolecules (5 distinct):