Filter by :
Entry Information
Entry status
(1)
Experimental methods
(4)
X-ray diffraction
(65)
Hybrid
(1)
Neutron Diffraction
(1)
Solution NMR
(1)
Authors
(146)
Sugiyama S
(54)
Matsuoka S
(53)
Murata M
(53)
Hayashi F
(43)
Inoue Y
(43)
Sonoyama M
(43)
Tsuchikawa H
(43)
Kakinouchi K
(34)
Nakano R
(14)
Matsuoka D
(10)
Hara T
(9)
Inoue T
(8)
Mizohata E
(8)
Murakami S
(7)
Hirose M
(6)
Ishida H
(6)
Ano H
(5)
Ichihara O
(5)
Kimura SR
(5)
Lethu S
(5)
Veerkamp JH
(5)
Niiyama M
(4)
Sacchettini JC
(4)
Scapin G
(4)
Takahashi J
(4)
Kromminga A
(3)
Patel SB
(3)
Young AC
(3)
Young ACM
(3)
Cousido-Siah A
(2)
Ehler A
(2)
Kuhn B
(2)
Mitschler A
(2)
Podjarny A
(2)
Rudolph MG
(2)
Wang L
(2)
Alvarez A
(1)
Alvarez HA
(1)
Awwa M
(1)
Benz J
(1)
Blakeley MP
(1)
Bogdan DM
(1)
Carbonetti GS
(1)
Carlevaro CM
(1)
Carlevaro M
(1)
Carney DF
(1)
Ceccarelli SM
(1)
Che J
(1)
Claudot J
(1)
Clement T
(1)
Conte A
(1)
Deutsch DG
(1)
Elmes MW
(1)
Espinosa Y
(1)
Espinosa YR
(1)
Fadel F
(1)
Falcone J
(1)
Freitag M
(1)
Gan Q
(1)
Gao L
(1)
Gasser R
(1)
Guillot B
(1)
Haertlein M
(1)
Hatsui T
(1)
Hoshina M
(1)
Howard E
(1)
Howard EI
(1)
Hsu HC
(1)
Hu K
(1)
Ihara K
(1)
Iwata S
(1)
Jing Y
(1)
Joseph OM
(1)
Joti Y
(1)
Kaczocha M
(1)
Kameshima T
(1)
Kuhne H
(1)
Kühne H
(1)
Li H
(1)
Li S
(1)
Li Y
(1)
Liu Y
(1)
Liu Yh
(1)
Lucke C
(1)
Luecke C
(1)
Lv P
(1)
Lücke C
(1)
Magnone C
(1)
Masuda T
(1)
Matsumura H
(1)
Moulin M
(1)
Myers M
(1)
Nango E
(1)
Neidhart W
(1)
Obst U
(1)
Obst-Sander U
(1)
Ojima I
(1)
Ottaviani G
(1)
Pan D
(1)
Park J
(1)
More...
Homo / hetero assembly
(1)
homo
(66)
Assembly composition
(1)
protein structure
(66)
Assembly polymer count
(1)
monomer
(66)
Resolution distribution
0.5 - 1
(42)
1.0 - 1.5
(15)
1.5 - 2
(4)
2.0 - 2.5
(3)
2.5 - 3
(1)
Release year distribution
1990 - 1995
(4)
1995 - 2000
(3)
2000 - 2005
(1)
2010 - 2015
(9)
2015 - 2020
(12)
2020 - 2025
(46)
Journal
(13)
To be published
(48)
Angew Chem Int Ed Engl
(5)
Structure
(3)
Biochem J
(1)
Bioorg Med Chem Lett
(1)
Eur J Med Chem
(1)
IUCrJ
(1)
J Biol Chem
(1)
J Phys Chem B
(1)
J Synchrotron Radiat
(1)
Life (Basel)
(1)
Nat Methods
(1)
Proteins
(1)
Macromolecules
Organism superkingdom
(1)
Eukaryota
(66)
Organism name
(2)
Homo sapiens
(65)
Mus musculus
(1)
Molecule name
(8)
Fatty acid-binding protein 3
(66)
Fatty acid-binding protein, heart
(66)
H-FABP
(66)
Heart-type fatty acid-binding protein
(66)
MDGI
(66)
Mammary-derived growth inhibitor
(66)
M-FABP
(65)
Muscle fatty acid-binding protein
(65)
Molecule type
(1)
Protein
(66)
Gene names
(5)
FABP11
(65)
FABP3
(65)
MDGI
(65)
Fabp3
(1)
Fabph1
(1)
Interacting ligands
(55)
P6G : HEXAETHYLENE GLYCOL
(33)
PLM : PALMITIC ACID
(8)
1PE : PENTAETHYLENE GLYCOL
(7)
GLP : 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose
(4)
OLA : OLEIC ACID
(4)
PG4 : TETRAETHYLENE GLYCOL
(4)
STE : STEARIC ACID
(4)
ACY : ACETIC ACID
(3)
2AN : 8-ANILINO-1-NAPHTHALENE SULFONATE
(2)
3IM : 2,6-ditert-butyl-4-methyl-phenol
(2)
6NA : HEXANOIC ACID
(2)
DKA : DECANOIC ACID
(2)
ELA : 9-OCTADECENOIC ACID
(2)
KNA : nonanoic acid
(2)
MYR : MYRISTIC ACID
(2)
SO4 : SULFATE ION
(2)
08O : (Z)-docos-13-enoic acid
(1)
11A : UNDECANOIC ACID
(1)
4EI : perfluoroheptanoic acid
(1)
4I1 : Petroselinic acid
(1)
4I6 : perfluorononanoic acid
(1)
4IF : cis-vaccenic acid
(1)
5M8 : 6-chloranyl-2-methyl-4-phenyl-quinoline-3-carboxylic acid
(1)
87K : henicosanoic acid
(1)
8PF : pentadecafluorooctanoic acid
(1)
ACD : ARACHIDONIC ACID
(1)
BZV : tetracosanoic acid
(1)
CL : CHLORIDE ION
(1)
DAO : LAURIC ACID
(1)
DCR : icosanoic acid
(1)
DKL : heptane-1,1-diol
(1)
EIC : LINOLEIC ACID
(1)
EO3 : docosanoic acid
(1)
EPA : 5,8,11,14,17-EICOSAPENTAENOIC ACID
(1)
EW8 : nonadecanoic acid
(1)
F15 : PENTADECANOIC ACID
(1)
F23 : TRICOSANOIC ACID
(1)
FLR : (2R)-2-(3-fluoro-4-phenyl-phenyl)propanoic acid
(1)
HA4 : 2-cyclohexadecylethanoic acid
(1)
HXA : DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID
(1)
IBP : IBUPROFEN
(1)
IMN : INDOMETHACIN
(1)
J1W : 2-Bromopalmitic acid
(1)
J3R : 3-[methyl-(4-nitro-2,1,3-benzoxadiazol-7-yl)amino]propanoic acid
(1)
JEC : 4-[2-[1-(4-bromophenyl)-5-phenyl-pyrazol-3-yl]phenoxy]butanoic acid
(1)
LNL : ALPHA-LINOLENIC ACID
(1)
NER : (15E)-TETRACOS-15-ENOIC ACID
(1)
NI : NICKEL (II) ION
(1)
OCA : OCTANOIC ACID (CAPRYLIC ACID)
(1)
PAM : PALMITOLEIC ACID
(1)
PEG : DI(HYDROXYETHYL)ETHER
(1)
SO3 : SULFITE ION
(1)
TDA : N-TRIDECANOIC ACID
(1)
VCA : VACCENIC ACID
(1)
X90 : heptadecanoic acid
(1)
Function and Biology
Biological function
(8)
cytoskeletal protein binding
(66)
fatty acid binding
(66)
icosatetraenoic acid binding
(66)
lipid binding
(66)
long-chain fatty acid binding
(66)
long-chain fatty acid transmembrane transporter activity
(66)
oleic acid binding
(66)
protein binding
(65)
Biological process
(14)
brown fat cell differentiation
(66)
cholesterol homeostasis
(66)
fatty acid metabolic process
(66)
intracellular lipid transport
(66)
long-chain fatty acid transport
(66)
phospholipid homeostasis
(66)
positive regulation of long-chain fatty acid import into cell
(66)
positive regulation of phospholipid biosynthetic process
(66)
regulation of fatty acid oxidation
(66)
regulation of phosphatidylcholine biosynthetic process
(66)
response to fatty acid
(66)
response to insulin
(66)
response to xenobiotic stimulus
(66)
negative regulation of cell population proliferation
(65)
Biological cell component
(6)
cytoplasm
(66)
cytosol
(66)
extracellular space
(66)
sarcoplasm
(66)
extracellular exosome
(65)
nucleus
(65)
Sequence and Structure classification
SCOP fold
(1)
Lipocalins
(5)
SCOP family
(1)
Fatty acid binding protein-like
(5)
CATH class
(1)
Mainly Beta
(20)
CATH topology
(1)
Lipocalin
(20)
Pfam accession / name
(1)
PF00061 : Lipocalin
(66)
Experimental Information
Diffraction protocol
(2)
Single wavelength
(61)
Laue
(1)
Diffraction radiation source type
(4)
Synchrotron
(59)
Free-electron laser
(1)
Nuclear reactor
(1)
Rotating anode
(1)
Diffraction source
(12)
SPRING-8 BEAMLINE BL44XU
(35)
SPRING-8 BEAMLINE BL38B1
(16)
SLS BEAMLINE X10SA
(2)
SPRING-8 BEAMLINE BL41XU
(2)
ALS BEAMLINE 5.0.1
(1)
APS BEAMLINE 31-ID
(1)
ILL BEAMLINE LADI III
(1)
PHOTON FACTORY BEAMLINE BL-17A
(1)
RIGAKU
(1)
SACLA BEAMLINE BL3
(1)
SLS BEAMLINE X06DA
(1)
SLS BEAMLINE X06SA
(1)
Synchrotron site
(7)
SPring-8
(52)
SLS
(4)
ALS
(1)
APS
(1)
ILL
(1)
Photon Factory
(1)
SACLA
(1)
Diffraction detector type
(3)
CCD
(54)
Pixel
(7)
Image plate
(1)
Refinement software
(5)
REFMAC
(54)
PHENIX
(7)
TNT
(4)
X-PLOR
(4)
SHELXL
(3)
NMR
NMR software packages
(4)
AURELIA
(1)
DYANA
(1)
Discover
(1)
XwinNMR
(1)
NMR Field Strength
500.0 - 600
(1)
600.0 - 700
(1)
Representative Structures
Representative Structures
Entries
Macromolecules
Compounds
Protein families
...
Entries 1 to 10 of 66
Select all entries on this page
The 0.88 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with alpha-llinolenic acid
Sugiyama S, Nakano R, Matsuoka S, Tsuchikawa H, Sonoyama M, Inoue Y, Hayashi F, Murata M
To be published
Source organism: Homo sapiens
Assembly composition: protein only structure
Assembly name:
Fatty acid-binding protein, heart
(Preferred)
search this complex
PDBe complex ID:
PDB-CPX-138660 (Preferred)
search this ID
X-ray structure of the human heart fatty acid-binding protein complexed with S-Ibuprofen
Sugiyama S, Kakinouchi K, Matsuoka S, Tsuchikawa H, Sonoyama M, Inoue Y, Hayashi F, Murata M
To be published
Source organism: Homo sapiens
Assembly composition: protein only structure
Assembly name:
Fatty acid-binding protein, heart
(Preferred)
search this complex
PDBe complex ID:
PDB-CPX-138660 (Preferred)
search this ID
X-ray structure of high-strength hydrogel-grown FABP3 crystal soaked in 50% DMSO solution containing 4-[2-[1-(4-bromophenyl)-5-phenyl-1H-pyrazol-3-yl]phenoxy] (HA174)
Sugiyama S, Kakinouchi K, Nakano R, Matsuoka S, Tsuchikawa H, Sonoyama M, Inoue Y, Hayashi F, Murata M
To be published
Source organism: Homo sapiens
Assembly composition: protein only structure
Assembly name:
Fatty acid-binding protein, heart
(Preferred)
search this complex
PDBe complex ID:
PDB-CPX-138660 (Preferred)
search this ID
X-ray structure of the human heart fatty acid-binding protein complexed with the fluorescent probe HA527
Takabayashi M, Yokota J, Matsuoka S, Tsuchikawa H, Sonoyama M, Inoue Y, Hayashi F, Sugiyama S
To be published
Source organism: Homo sapiens
Assembly composition: protein only structure
Assembly name:
Fatty acid-binding protein, heart
(Preferred)
search this complex
PDBe complex ID:
PDB-CPX-138660 (Preferred)
search this ID
The 1.28A structure of human FABP3 F16V mutant complexed with palmitic acid at room temperature
Matsuoka D, Sugiyama S, Kakinouchi K, Niiyama M, Murata M, Matsuoka S
To be published
Source organism: Homo sapiens
Assembly composition: protein only structure
Bound ligands:
PLM
Assembly name:
Fatty acid-binding protein, heart
(Preferred)
search this complex
PDBe complex ID:
PDB-CPX-138660 (Preferred)
search this ID
High Resolution X-Ray and Neutron diffraction structure of H-FABP
Podjarny AD, Howard EI, Blakeley MP, Guillot B
IUCrJ
(2016)
[PMID: 27006775 ]
Source organism: Homo sapiens
Assembly composition: protein only structure
Assembly name:
Fatty acid-binding protein, heart
(Preferred)
search this complex
PDBe complex ID:
PDB-CPX-138660 (Preferred)
search this ID
X-ray diffraction, Neutron Diffraction, Hybrid
0.98Å resolution
Released: 9 Mar 2016
DOI:
10.2210/pdb5ce4
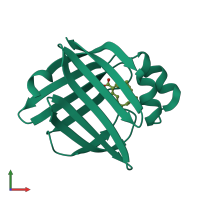

The 1.02A structure of human FABP3 M20S mutant complexed with palmitic acid
Matsuoka D, Sugiyama S, Kakinouchi K, Niiyama M, Murata M, Matsuoka S
To be published
Source organism: Homo sapiens
Assembly composition: protein only structure
Assembly name:
Fatty acid-binding protein, heart
(Preferred)
search this complex
PDBe complex ID:
PDB-CPX-138660 (Preferred)
search this ID
The 0.90A structure of human FABP3 F16V mutant complexed with palmitic acid
Matsuoka D, Sugiyama S, Kakinouchi K, Niiyama M, Murata M, Matsuoka S
To be published
Source organism: Homo sapiens
Assembly composition: protein only structure
Assembly name:
Fatty acid-binding protein, heart
(Preferred)
search this complex
PDBe complex ID:
PDB-CPX-138660 (Preferred)
search this ID
The 0.93 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with tridecanoic acid
Sugiyama S, Takahashi J, Matsuoka S, Tsuchikawa H, Sonoyama M, Inoue Y, Hayashi F, Murata M
To be published
Source organism: Homo sapiens
Assembly composition: protein only structure
Assembly name:
Fatty acid-binding protein, heart
(Preferred)
search this complex
PDBe complex ID:
PDB-CPX-138660 (Preferred)
search this ID
human FABP3 in complex with 6-Chloro-2-methyl-4-phenyl-quinoline-3-carboxylic acid
Ehler A, Rudolph MG
Bioorg Med Chem Lett
(2016)
[PMID: 27658368 ]
Source organism: Homo sapiens
Assembly composition: protein only structure
Assembly name:
Fatty acid-binding protein, heart
(Preferred)
search this complex
PDBe complex ID:
PDB-CPX-138660 (Preferred)
search this ID
...
Entries 1 to 10 of 66




























