About
Sequence Logos and its variants are the most commonly used method for visualisation of multiple sequence alignments (MSAs) and sequence motifs. They provide consensus-based summaries of the sequences in the alignment. Consequently, sequences cannot be identified anymore in the visualisation and covariant sites are not easily detectable. We recently proposed Sequence Bundles[1], a motif visualisation technique that retains sequence identity and maintains a one-to-one relationship between sequences and their graphical representation.
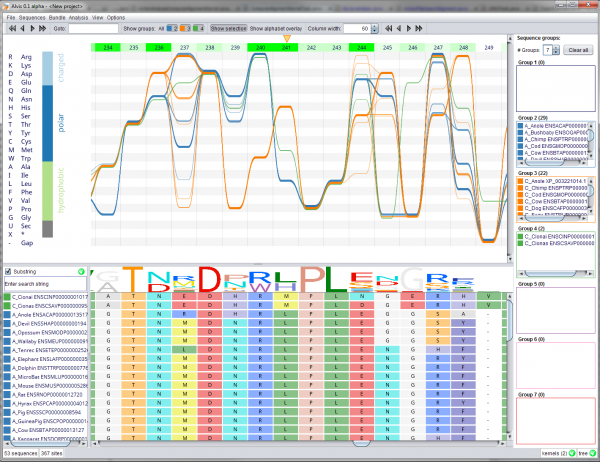
Alvis is an open-source platform for the joint explorative analysis of MSAs and phylogenetic trees, employing Sequence Bundles as its main visualisation method. Alvis combines the power of the visualisation method with an interactive toolkit allowing detection of covariant sites, annotation of trees with synapomorphies and homoplasies, and motif detection. It also offers numerical analysis functionality, such as dimension reduction and classification. Alvis is user-friendly, highly customisable, and can export results in publication-quality figures.
Sequence Bundles and Alvis were devised in a collaboration between the Goldman Group at EBI and Science Practice, a design and research company in London.
Jump to: Screenshot Tutorial Download Web Application References
Screenshot
Tutorial
We have recorded a video tutorial illustrating some of the functionality available in Alvis.
Download
The standalone version of Alvis is available from Bitbucket.
Web Application
A simplified version of Alvis is also available as a web application.
References
[1]Kultys, M., Nicholas, L., Schwarz, R., Goldman, N., & King, J. (2014). Sequence Bundles: a novel method for visualising, discovering and exploring sequence motifs. BMC Proceedings, 8(Suppl 2), S8. http://doi.org/10.1186/1753-6561-8-S2-S8