Quinoprotein glucose dehydrogenase
The soluble quinoprotein glucose dehydrogenase (s-GDH, EC 1.1.99.17) from Acinetobacter calcoaceticus is a classical quinoprotein which requires the cofactor pyrroloquinoline quinone (PQQ) to oxidise glucose to gluconolactone with the concomitant reduction of ubiquinone.
The reaction mechanism of PQQ-dependent enzymes has remained controversial due to the absence of comprehensive structural data. Three alternative proposals have been suggested: the addition-elimination reaction in which the glucose becomes covalently attached to the PQQ, the hydride transfer to the C5 of the PQQ, and the hydride transfer to the C4 of the PQQ [PMID:10508152, PMID:12686124].
Reference Protein and Structure
- Sequence
-
P13650
 (1.1.5.2)
(1.1.5.2)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Acinetobacter calcoaceticus (Bacteria)

- PDB
-
1c9u
- CRYSTAL STRUCTURE OF THE SOLUBLE QUINOPROTEIN GLUCOSE DEHYDROGENASE IN COMPLEX WITH PQQ
(2.2 Å)



- Catalytic CATH Domains
-
2.120.10.30
 (see all for 1c9u)
(see all for 1c9u)
- Cofactors
- Pyrroloquinoline quinone(3-) (1), Calcium(2+) (1), Coenzyme q10 (1) Metal MACiE
Enzyme Reaction (EC:1.1.5.2)
Enzyme Mechanism
Introduction
Crystallographic evidence [PMID:10508152] strongly supports the direct hydride transfer from the glucose to the C5 of the PQQ cofactor. Followed by re-oxidation of the PQQ cofactor by ubiquinone.
Catalytic Residues Roles
| UniProt | PDB* (1c9u) | ||
| Arg252 | Arg228A | Stabilises the negatively charged intermediates that are formed on the PQQ cofactor during the course of the reaction. | hydrogen bond donor, electrostatic stabiliser |
| His168 | His144A | Acts as a general acid/base. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Asp187 | Asp163A | Activates Hiss144 to better function as a general acid/base. | activator, hydrogen bond acceptor, electrostatic stabiliser |
| Ala293 (main-C), Tyr295 (main-C), Asp297, Glu333 | Ala269A (main-C), Tyr271A (main-C), Asp273A, Glu309A | Forms part of the catalytic calcium binding site | metal ligand |
Chemical Components
bimolecular elimination, hydride transfer, aromatic bimolecular nucleophilic addition, overall reactant used, cofactor used, intermediate formation, overall product formed, proton transfer, assisted keto-enol tautomerisation, rate-determining step, bimolecular nucleophilic addition, native state of cofactor regenerated, native state of cofactor is not regenerated, intermediate terminated, native state of enzyme regeneratedReferences
- Dewanti AR et al. (2000), Biochemistry, 39, 9384-9392. Ca2+-Assisted, Direct Hydride Transfer, and Rate-Determining Tautomerization of C5-Reduced PQQ to PQQH2, in the Oxidation ofβ-d-Glucose by Soluble, Quinoprotein Glucose Dehydrogenase. DOI:10.1021/bi992810x. PMID:10924133.
- Leopoldini M et al. (2007), Chemistry, 13, 2109-2117. The Preferred Reaction Path for the Oxidation of Methanol by PQQ-Containing Methanol Dehydrogenase: Addition–Elimination versus Hydride-Transfer Mechanism. DOI:10.1002/chem.200601123. PMID:17149777.
- Reddy SY et al. (2004), Protein Sci, 13, 1965-1978. Determination of enzyme mechanisms by molecular dynamics: Studies on quinoproteins, methanol dehydrogenase, and soluble glucose dehydrogenase. DOI:10.1110/ps.04673404. PMID:15273299.
- Oubrie A (2003), Biochim Biophys Acta, 1647, 143-151. Structure and mechanism of soluble glucose dehydrogenase and other PQQ-dependent enzymes. DOI:10.1016/s1570-9639(03)00087-6. PMID:12686124.
- Zheng YJ et al. (2001), Proc Natl Acad Sci U S A, 98, 432-434. Catalytic mechanism of quinoprotein methanol dehydrogenase: A theoretical and x-ray crystallographic investigation. DOI:10.1073/pnas.021547498. PMID:11149955.
- Jongejan A et al. (2001), J Comput Chem, 22, 1732-1749. Direct hydride transfer in the reaction mechanism of quinoprotein alcohol dehydrogenases: a quantum mechanical investigation. DOI:10.1002/jcc.1128. PMID:12116408.
- Oubrie A et al. (1999), EMBO J, 18, 5187-5194. Structure and mechanism of soluble quinoprotein glucose dehydrogenase. DOI:10.1093/emboj/18.19.5187. PMID:10508152.
- Olsthoorn AJ et al. (1998), Biochemistry, 37, 13854-13861. On the Mechanism and Specificity of Soluble, Quinoprotein Glucose Dehydrogenase in the Oxidation of Aldose Sugars†. DOI:10.1021/bi9808868. PMID:9753475.

Step 1. His144 deprotonates the C1-OH of glucose, initiating the elimination of a hydride, which is added to the C5 of the PQQ cofactor.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His144A | hydrogen bond acceptor, hydrogen bond donor |
| Asp163A | hydrogen bond acceptor, activator |
| Arg228A | hydrogen bond donor |
| Asp273A | metal ligand |
| Glu309A | metal ligand |
| Ala269A (main-C) | metal ligand |
| Tyr271A (main-C) | metal ligand |
| His144A | proton acceptor |
Chemical Components
ingold: bimolecular elimination, hydride transfer, ingold: aromatic bimolecular nucleophilic addition, overall reactant used, cofactor used, intermediate formation, overall product formedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His144A | hydrogen bond donor |
| Asp163A | hydrogen bond acceptor, electrostatic stabiliser |
| Arg228A | hydrogen bond donor, electrostatic stabiliser |
| Asp273A | metal ligand |
| Glu309A | metal ligand |
| Ala269A (main-C) | metal ligand |
| Tyr271A (main-C) | metal ligand |
| His144A | proton donor |
Chemical Components
proton transfer, intermediate formation
Step 3. His144 deprotonates the C5 carbon of PQQ, forming the C4-oxyanion.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His144A | hydrogen bond acceptor, hydrogen bond donor |
| Asp163A | hydrogen bond acceptor, activator |
| Arg228A | hydrogen bond donor |
| Asp273A | metal ligand |
| Glu309A | metal ligand |
| Ala269A (main-C) | metal ligand |
| Tyr271A (main-C) | metal ligand |
| His144A | proton acceptor |
Chemical Components
proton transfer, assisted keto-enol tautomerisation, intermediate formation, rate-determining stepCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His144A | hydrogen bond donor |
| Asp163A | hydrogen bond acceptor, electrostatic stabiliser |
| Arg228A | hydrogen bond donor, electrostatic stabiliser |
| Asp273A | metal ligand |
| Glu309A | metal ligand |
| Ala269A (main-C) | metal ligand |
| Tyr271A (main-C) | metal ligand |
| His144A | proton donor |
Chemical Components
proton transfer, intermediate formation
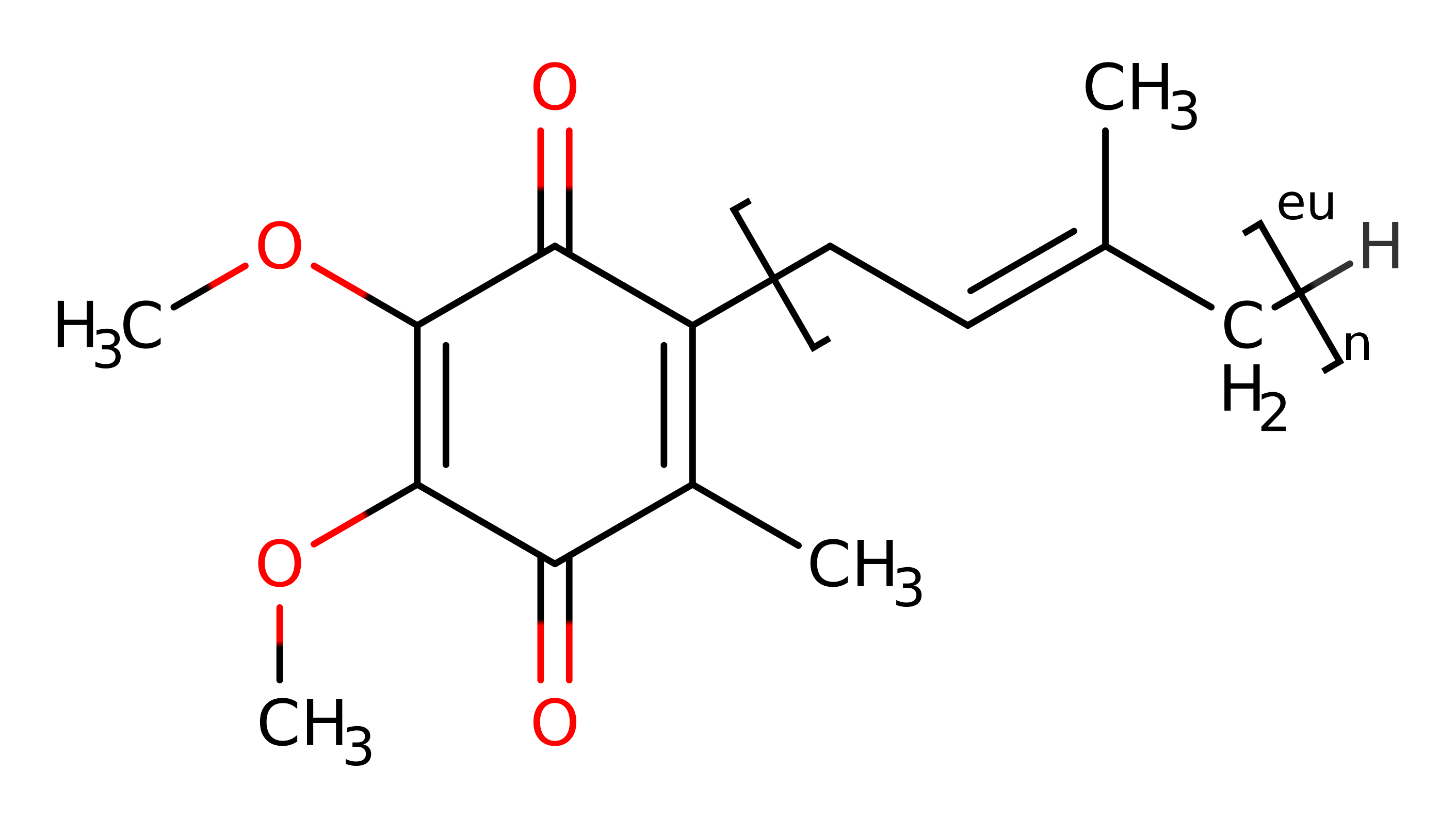
Step 5. The quinone re-oxidises PQQ, the exact mechanism is unclear. The PQQ cofactor can be re-oxidised in a single two-electron transfer step. Alternatively, re-oxidation can be achieved in two separate one-electron transfer steps via the free radical semiquinone form.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His144A | hydrogen bond donor |
| Asp163A | hydrogen bond acceptor |
| Arg228A | hydrogen bond donor |
| Asp273A | metal ligand |
| Glu309A | metal ligand |
| Ala269A (main-C) | metal ligand |
| Tyr271A (main-C) | metal ligand |
Chemical Components
ingold: bimolecular elimination, hydride transfer, ingold: bimolecular nucleophilic addition, overall reactant used, native state of cofactor regenerated, cofactor used, native state of cofactor is not regenerated, intermediate terminated, overall product formed, native state of enzyme regeneratedIntroduction
Alternative mechanism in which a direct hydride transfer from the glucose to the C4 of the PQQ cofactor occurs. Followed by re-oxidation of the PQQ cofactor by ubiquinone.
Catalytic Residues Roles
| UniProt | PDB* (1c9u) | ||
| Arg252 | Arg228A | Stabilises the negatively charged intermediates that are formed on the PQQ cofactor during the course of the reaction. | hydrogen bond donor, electrostatic stabiliser |
| His168 | His144A | Acts as a general acid/base. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Asp187 | Asp163A | Activates Hiss144 to better function as a general acid/base. | activator, hydrogen bond acceptor, electrostatic stabiliser |
| Ala293 (main-C), Tyr295 (main-C), Asp297, Glu333 | Ala269A (main-C), Tyr271A (main-C), Asp273A, Glu309A | Forms part of the catalytic calcium binding site. | metal ligand |
Chemical Components
bimolecular elimination, hydride transfer, aromatic bimolecular nucleophilic addition, overall reactant used, cofactor used, intermediate formation, overall product formed, proton transfer, assisted keto-enol tautomerisation, bimolecular nucleophilic addition, native state of cofactor regenerated, native state of cofactor is not regenerated, intermediate terminated, native state of enzyme regeneratedReferences
- Zheng YJ et al. (1997), Proc Natl Acad Sci U S A, 94, 11881-11886. Conformation of coenzyme pyrroloquinoline quinone and role of Ca2+ in the catalytic mechanism of quinoprotein methanol dehydrogenase. PMID:9342331.
- Oubrie A et al. (1999), EMBO J, 18, 5187-5194. Structure and mechanism of soluble quinoprotein glucose dehydrogenase. DOI:10.1093/emboj/18.19.5187. PMID:10508152.

Step 1. His144 deprotonates the C1-OH of glucose, initiating the elimination of a hydride, which is added to the C4 of the PQQ cofactor.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr271A (main-C) | metal ligand |
| Ala269A (main-C) | metal ligand |
| Glu309A | metal ligand |
| Asp273A | metal ligand |
| His144A | hydrogen bond acceptor, hydrogen bond donor |
| Asp163A | hydrogen bond acceptor, activator |
| Arg228A | hydrogen bond donor |
| His144A | proton acceptor |
Chemical Components
ingold: bimolecular elimination, hydride transfer, ingold: aromatic bimolecular nucleophilic addition, overall reactant used, cofactor used, intermediate formation, overall product formedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp273A | metal ligand |
| Glu309A | metal ligand |
| Ala269A (main-C) | metal ligand |
| Tyr271A (main-C) | metal ligand |
| Asp163A | activator |
| Arg228A | electrostatic stabiliser |
| His144A | proton donor |
Chemical Components
proton transfer
Step 3. His144 deprotonates the C4 carbon of PQQ, forming the C5-oxyanion.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp273A | metal ligand |
| Glu309A | metal ligand |
| Ala269A (main-C) | metal ligand |
| Tyr271A (main-C) | metal ligand |
| Asp163A | electrostatic stabiliser |
| Arg228A | electrostatic stabiliser |
| His144A | proton acceptor |
Chemical Components
proton transfer, assisted keto-enol tautomerisationCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr271A (main-C) | metal ligand |
| Ala269A (main-C) | metal ligand |
| Glu309A | metal ligand |
| Asp273A | metal ligand |
| His144A | hydrogen bond donor |
| Asp163A | hydrogen bond acceptor, electrostatic stabiliser |
| Arg228A | hydrogen bond donor, electrostatic stabiliser |
| His144A | proton donor |
Chemical Components
proton transfer, intermediate formation
Step 5. The quinone re-oxidises PQQ, the exact mechanism is unclear. The PQQ cofactor can be re-oxidised in a single two-electron transfer step. Alternatively, re-oxidation can be achieved in two separate one-electron transfer steps via the free radical semiquinone form.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr271A (main-C) | metal ligand |
| Ala269A (main-C) | metal ligand |
| Glu309A | metal ligand |
| Asp273A | metal ligand |
| His144A | hydrogen bond donor |
| Asp163A | hydrogen bond acceptor |
| Arg228A | hydrogen bond donor |
Chemical Components
ingold: bimolecular elimination, hydride transfer, ingold: bimolecular nucleophilic addition, overall reactant used, native state of cofactor regenerated, cofactor used, native state of cofactor is not regenerated, intermediate terminated, overall product formed, native state of enzyme regeneratedIntroduction
The addition-elimination reaction comprises general base catalysed proton abstraction, followed by the formation of a covalent PQQ-substrate adduct, and oxidative elimination of the product. Followed by re-oxidation of the PQQ cofactor by ubiquinone.
Catalytic Residues Roles
| UniProt | PDB* (1c9u) | ||
| Arg252 | Arg228A | Stabilises the negatively charged intermediates that are formed on the PQQ cofactor during the course of the reaction. | hydrogen bond donor, electrostatic stabiliser |
| His168 | His144A | Acts as a general acid/base. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Asp187 | Asp163A | Activates Hiss144 to better function as a general acid/base. | activator, hydrogen bond acceptor, electrostatic stabiliser |
| Ala293 (main-C), Tyr295 (main-C), Asp297, Glu333 | Ala269A (main-C), Tyr271A (main-C), Asp273A, Glu309A | Forms part of the catalytic calcium binding site. | metal ligand |
Chemical Components
bimolecular elimination, hydride transfer, aromatic bimolecular nucleophilic addition, overall reactant used, cofactor used, intermediate formation, overall product formed, proton transfer, intramolecular elimination, bimolecular nucleophilic addition, native state of cofactor regenerated, native state of cofactor is not regenerated, intermediate terminated, native state of enzyme regeneratedReferences
- Itoh S et al. (1998), Biochemistry, 37, 6562-6571. Model Studies on Calcium-Containing Quinoprotein Alcohol Dehydrogenases. Catalytic Role of Ca2+for the Oxidation of Alcohols by Coenzyme PQQ (4,5-Dihydro-4,5-dioxo-1H-pyrrolo[2,3-f]quinoline-2,7,9-tricarboxylic Acid)†. DOI:10.1021/bi9800092. PMID:9572874.
- Leopoldini M et al. (2007), Chemistry, 13, 2109-2117. The Preferred Reaction Path for the Oxidation of Methanol by PQQ-Containing Methanol Dehydrogenase: Addition–Elimination versus Hydride-Transfer Mechanism. DOI:10.1002/chem.200601123. PMID:17149777.
- Oubrie A et al. (2000), Protein Sci, 9, 1265-1273. Structural requirements of pyrroloquinoline quinone dependent enzymatic reactions. DOI:10.1110/ps.9.7.1265. PMID:10933491.
- Oubrie A et al. (1999), EMBO J, 18, 5187-5194. Structure and mechanism of soluble quinoprotein glucose dehydrogenase. DOI:10.1093/emboj/18.19.5187. PMID:10508152.
- Olsthoorn AJ et al. (1998), Biochemistry, 37, 13854-13861. On the Mechanism and Specificity of Soluble, Quinoprotein Glucose Dehydrogenase in the Oxidation of Aldose Sugars†. DOI:10.1021/bi9808868. PMID:9753475.
- Itoh S et al. (1997), J Am Chem Soc, 119, 439-440. Modeling of the Chemistry of Quinoprotein Methanol Dehydrogenase. Oxidation of Methanol by Calcium Complex of Coenzyme PQQ via Addition−Elimination Mechanism. DOI:10.1021/ja963366d.
- Frank J Jr et al. (1989), Eur J Biochem, 184, 187-195. On the mechanism of inhibition of methanol dehydrogenase by cyclopropane-derived inhibitors. PMID:2550226.

Step 1. His144 deprotonates the C1-OH of glucose, initiating the addition reaction of the glucose substrate to the the C5 of the PQQ cofactor.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His144A | hydrogen bond acceptor, hydrogen bond donor |
| Asp163A | hydrogen bond acceptor, activator |
| Arg228A | hydrogen bond donor |
| Asp273A | metal ligand |
| Glu309A | metal ligand |
| Ala269A (main-C) | metal ligand |
| Tyr271A (main-C) | metal ligand |
| His144A | proton acceptor |
Chemical Components
ingold: bimolecular elimination, hydride transfer, ingold: aromatic bimolecular nucleophilic addition, overall reactant used, cofactor used, intermediate formation, overall product formedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp273A | metal ligand |
| Glu309A | metal ligand |
| Ala269A (main-C) | metal ligand |
| Tyr271A (main-C) | metal ligand |
| His144A | proton donor |
Chemical Components
proton transfer
Step 3. The PQQ abstracts a proton from the covalently bound adduct, eliminating it as the aldehyde product.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp273A | metal ligand |
| Glu309A | metal ligand |
| Ala269A (main-C) | metal ligand |
| Tyr271A (main-C) | metal ligand |
| Asp163A | electrostatic stabiliser |
| Arg228A | electrostatic stabiliser |
Chemical Components
ingold: intramolecular elimination, proton transfer
Step 4. The quinone re-oxidises PQQ, the exact mechanism is unclear. The PQQ cofactor can be re-oxidised in a single two-electron transfer step. Alternatively, re-oxidation can be achieved in two separate one-electron transfer steps via the free radical semiquinone form.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr271A (main-C) | metal ligand |
| Ala269A (main-C) | metal ligand |
| Glu309A | metal ligand |
| Asp273A | metal ligand |
| His144A | hydrogen bond donor |
| Asp163A | hydrogen bond acceptor |
| Arg228A | hydrogen bond donor |





 Download:
Download: 

 Download:
Download: 
 Download:
Download: