Aerobic carbon monoxide dehydrogenase
Carbon monoxide dehydrogenase (CODH), a member of the xanthine oxidase family based on its overall amino acid sequence and three-dimensional structure, catalyses the reversible oxidation of carbon monoxide to carbon dioxide with the loss of two electrons to a variety of electron acceptors can be used by these enzymes including ferredoxin, methyl viologen and benzyl viologen. The CO2 that is produced is assimilated by the Calvin-Benson-Basham cycle, while the electrons are transferred to a quinone via the FAD site, and continue through the electron transfer chain to a dioxygen terminal acceptor.
Under aerobic conditions carbon monoxide can be used as sole carbon and energy source by the carboxidotrophic bacteria, a taxonomically diverse group of obligate or facultative chemolithoautotrophic species. The aerobic enzymes (this entry) categorised so far are molybdenum-containing iron-sulphur proteins whilst the anerobic dehydrogenases are mostly nickel-containing iron-sulphur enzymes.
CODH is a dimer of heterotrimers, where each heterotrimer consists of a small iron-sulphur subunit, a medium FAD-containing flavoprotein subunit and a large molybdopterin-containign subunit which contains the active site [PMID:10430865, PMID:12475995]. The large subunit is divided into two domains. The N-terminal domain is composed of mainly of a five-and four-stranded mixed beta-sheet and interacts with the small and medium subunits. The C-terminal domain contains a region which interacts with its counterpart in the overall dimer, providing the main dimer contact, and a beta-sheet surrounded by alpha helices. The active site located in the large subunit and contains a novel dinuclear heterometal [CuSMo(=O)OH] cluster.
Reference Protein and Structure
- Sequences
-
P19921
 (1.2.5.3)
(1.2.5.3)
P19919 (1.2.5.3)
(1.2.5.3)
P19920 (1.2.5.3)
(1.2.5.3)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Oligotropha carboxidovorans OM5 (Bacteria)

- PDB
-
1n62
- Crystal Structure of the Mo,Cu-CO Dehydrogenase (CODH), n-butylisocyanide-bound state
(1.09 Å)



- Catalytic CATH Domains
-
3.30.365.10
 (see all for 1n62)
(see all for 1n62)
- Cofactors
- Cu(i)-s-mo(iv)(=o)o-nbic cluster (1), Di-mu-sulfido-diiron(2+) (2), Fadh2(2-) (1) Metal MACiE
Enzyme Reaction (EC:1.2.5.3)
Enzyme Mechanism
Introduction
In this proposal, the reaction proceeds via a stable C-S intermediate.
This mechanism starts in the oxidized Mo(VI)-Cu(I) state. The substrate carbon monoxide (CO) reaches the cluster through the substrate channel. CO coordinates to the copper (I) centre. This activates the CO and allows for the formation of the C-O bond between CO and the oxygen derived ligand on Mo(VI). This results in the reduction of the molybdenum to Mo(IV) and the formation of a five membered ring. Carbon dioxide is then released, and the molybdenum becomes a 4-coordinated Mo(IV). Reoxidation of the Mo(IV) centre by an external electron acceptor completes the proposed catalytic cycle. Electrons released are transferred via the sulfido ligand to yield the reduced Mo(IV) state. Cu(I) does not change oxidation state during the mechanism. The chain of redox active cofactors (type-I [2Fe2S] cluster, type-II [2Fe2S] cluster, and FAD) facilitates the reoxidation of the reduced Mo-ion through electron transfer from the active site Mo to external redox partners like cytochrome b561.
Catalytic Residues Roles
| UniProt | PDB* (1n62) | ||
| Glu763 | Glu763B | Glu-763 interacts through its carboxyl atom with the Mo ion in trans position to the oxo-group and is linked through hydrogen bonds to the active-site loop carrying Cys-388. This residue acts as a general acid/base in the reaction. | proton relay, proton acceptor, proton donor |
| Gln240 | Gln240B | The Neta2 group of Gln-240 is within 2.90 Angstroms hydrogen-bonding distance to the apical oxo-ligand at the Mo ion, helping to activate and stabilise the ligand. | electrostatic stabiliser |
| Cys388 | Cys388B | One of the copper ligands. Activates the copper centre. | metal ligand |
| Arg387 | Arg387B | Helps stabilise the negative charge in the active site. | electrostatic stabiliser |
Chemical Components
intramolecular nucleophilic addition, electron transfer, intermediate formation, cyclisation, intramolecular nucleophilic substitution, decoordination from a metal ion, coordination to a metal ion, proton transfer, overall reactant used, electron relay, cofactor used, native state of cofactor regenerated, inferred reaction step, intermediate terminated, overall product formed, proton relay, native state of enzyme regeneratedReferences
- Siegbahn PE et al. (2005), J Comput Chem, 26, 888-898. Quantum chemical modeling of CO oxidation by the active site of molybdenum CO dehydrogenase. DOI:10.1002/jcc.20230. PMID:15834924.
- Stein BW et al. (2014), Chem Commun (Camb), 50, 1104-1106. Orbital contributions to CO oxidation in Mo-Cu carbon monoxide dehydrogenase. DOI:10.1039/c3cc47705c. PMID:24322538.
- Pelzmann AM et al. (2014), J Biol Inorg Chem, 19, 1399-1414. Insights into the posttranslational assembly of the Mo-, S- and Cu-containing cluster in the active site of CO dehydrogenase of Oligotropha carboxidovorans. DOI:10.1007/s00775-014-1201-y. PMID:25377894.
- Shanmugam M et al. (2013), J Am Chem Soc, 135, 17775-17782. 13C and63,65Cu ENDOR studies of CO Dehydrogenase fromOligotropha carboxidovorans. Experimental Evidence in Support of a Copper–Carbonyl Intermediate. DOI:10.1021/ja406136f. PMID:24147852.
- Wilcoxen J et al. (2011), J Am Chem Soc, 133, 12934-12936. Substitution of Silver for Copper in the Binuclear Mo/Cu Center of Carbon Monoxide Dehydrogenase from Oligotropha carboxidovorans. DOI:10.1021/ja205073j. PMID:21774528.
- Wilcoxen J et al. (2011), Biochemistry, 50, 1910-1916. Reaction of the Molybdenum- and Copper-Containing Carbon Monoxide Dehydrogenase fromOligotropha carboxydovoranswith Quinones. DOI:10.1021/bi1017182. PMID:21275368.
- Zhang B et al. (2010), J Biol Chem, 285, 12571-12578. Kinetic and Spectroscopic Studies of the Molybdenum-Copper CO Dehydrogenase from Oligotropha carboxidovorans. DOI:10.1074/jbc.m109.076851. PMID:20178978.
- Hofmann M et al. (2005), J Biol Inorg Chem, 10, 490-495. The mechanism of Mo-/Cu-dependent CO dehydrogenase. DOI:10.1007/s00775-005-0661-5. PMID:15971074.
- Dobbek H et al. (2002), Proc Natl Acad Sci U S A, 99, 15971-15976. Catalysis at a dinuclear [CuSMo(O)OH] cluster in a CO dehydrogenase resolved at 1.1-A resolution. DOI:10.1073/pnas.212640899. PMID:12475995.
- Gremer L et al. (2000), J Biol Chem, 275, 1864-1872. Binding of Flavin Adenine Dinucleotide to Molybdenum-containing Carbon Monoxide Dehydrogenase from Oligotropha carboxidovorans: STRUCTURAL AND FUNCTIONAL ANALYSIS OF A CARBON MONOXIDE DEHYDROGENASE SPECIES IN WHICH THE NATIVE FLAVOPROTEIN HAS BEEN REPLACED BY ITS RECOMBINANT COUNTERPART PRODUCED IN ESCHERICHIA COLI. DOI:10.1074/jbc.275.3.1864. PMID:10636886.
- Meyer O et al. (2000), Biol Chem, 381, 865-876. The Role of Se, Mo and Fe in the Structure and Function of Carbon Monoxide Dehydrogenase. DOI:10.1515/bc.2000.108. PMID:11076018.
- Dobbek H et al. (1999), Proc Natl Acad Sci U S A, 96, 8884-8889. Crystal structure and mechanism of CO dehydrogenase, a molybdo iron-sulfur flavoprotein containing S-selanylcysteine. DOI:10.1073/pnas.96.16.8884. PMID:10430865.

Step 1. The Carbanion of carbon monoxide initiates a nucleophilic attack on the oxo group coordinated to the Mo(VI) centre, which results in the reduction of the molybdenum to Mo(IV). The C#O triple bond reduces in order to C=O.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys388B | metal ligand |
| Gln240B | electrostatic stabiliser |
| Arg387B | electrostatic stabiliser |
Chemical Components
ingold: intramolecular nucleophilic addition, electron transfer, intermediate formation, cyclisation
Step 2. The sulfur dianion bridging the Mo(IV) and Cu(I) centres initiates a nucleophilic attack on the carbocation in a substitution reaction that displaces the carbon from the Cu(I) centre.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys388B | metal ligand |
| Gln240B | electrostatic stabiliser |
| Arg387B | electrostatic stabiliser |
Chemical Components
ingold: intramolecular nucleophilic substitution, decoordination from a metal ion, intermediate formation
Step 3. The carbonyl oxygen initiates a nucleophilic attack on the Cu(I) centre in a substitution reaction that displaces the bridging sulfur ligand from the Cu(I) centre.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys388B | metal ligand |
| Gln240B | electrostatic stabiliser |
| Arg387B | electrostatic stabiliser |
Chemical Components
ingold: intramolecular nucleophilic substitution, coordination to a metal ion, decoordination from a metal ion, intermediate formation
Step 4. Glu763 deprotonates water, initiates a nucleophilic attack on the Mo(IV) centre, displacing the negatively charged oxygen of the carboxylate group in a substitution reaction, which initiates an elimination of the sulfur group, to reform the bridging sulfur dianion and the formation of carbon dioxide, which decoordinates from the metal centre.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu763B | proton acceptor |
Chemical Components
proton transfer, electron transfer, overall reactant used, electron relay, intermediate formation, cofactor used, native state of cofactor regenerated, inferred reaction step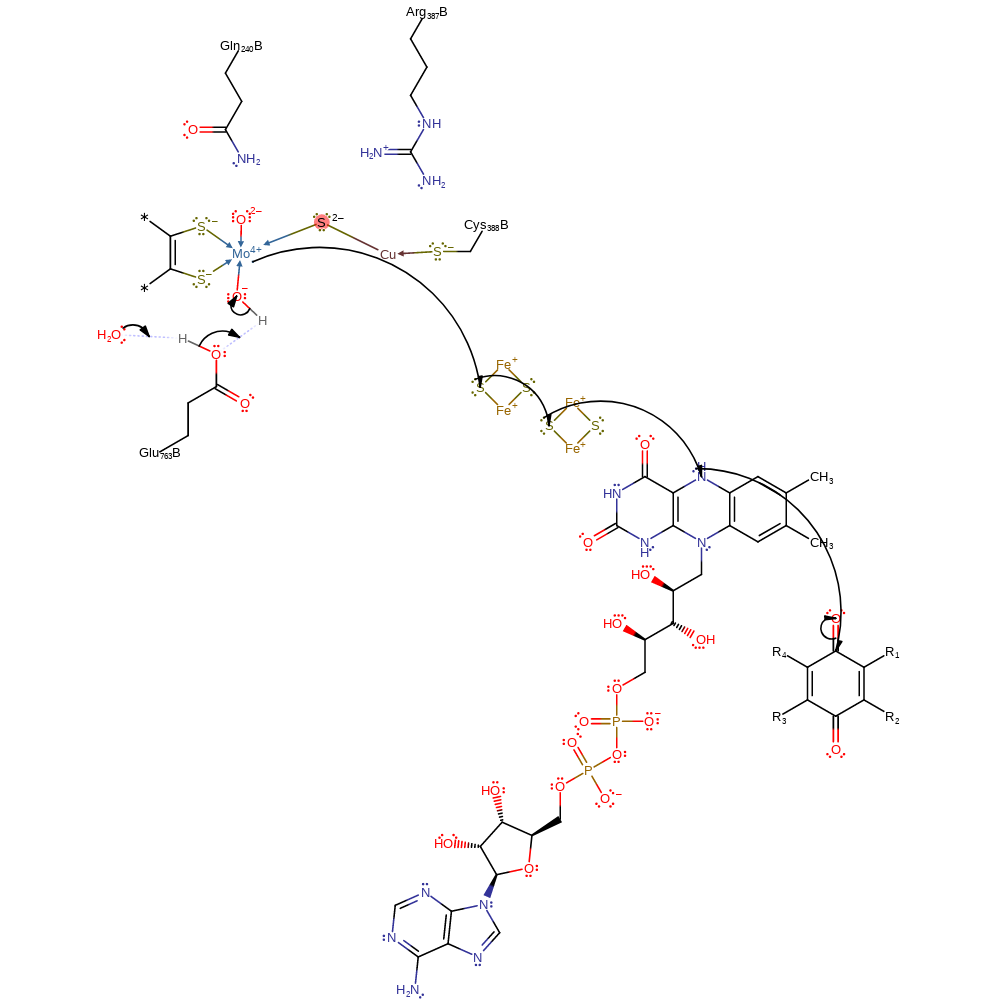
Step 5. Water deprotonates Glu763, which then deprotonates the Mo(V) bound hydroxide, initiating the second single electron transfer, through two iron-sulfur clusters to FAD, and thence to an external electron acceptor. This regenerates the cofactor.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys388B | metal ligand |
| Gln240B | electrostatic stabiliser |
| Arg387B | electrostatic stabiliser |
| Glu763B | proton acceptor, proton donor, proton relay |
Chemical Components
proton transfer, electron transfer, intermediate terminated, cofactor used, native state of cofactor regenerated, overall product formed, proton relay, electron relay, inferred reaction step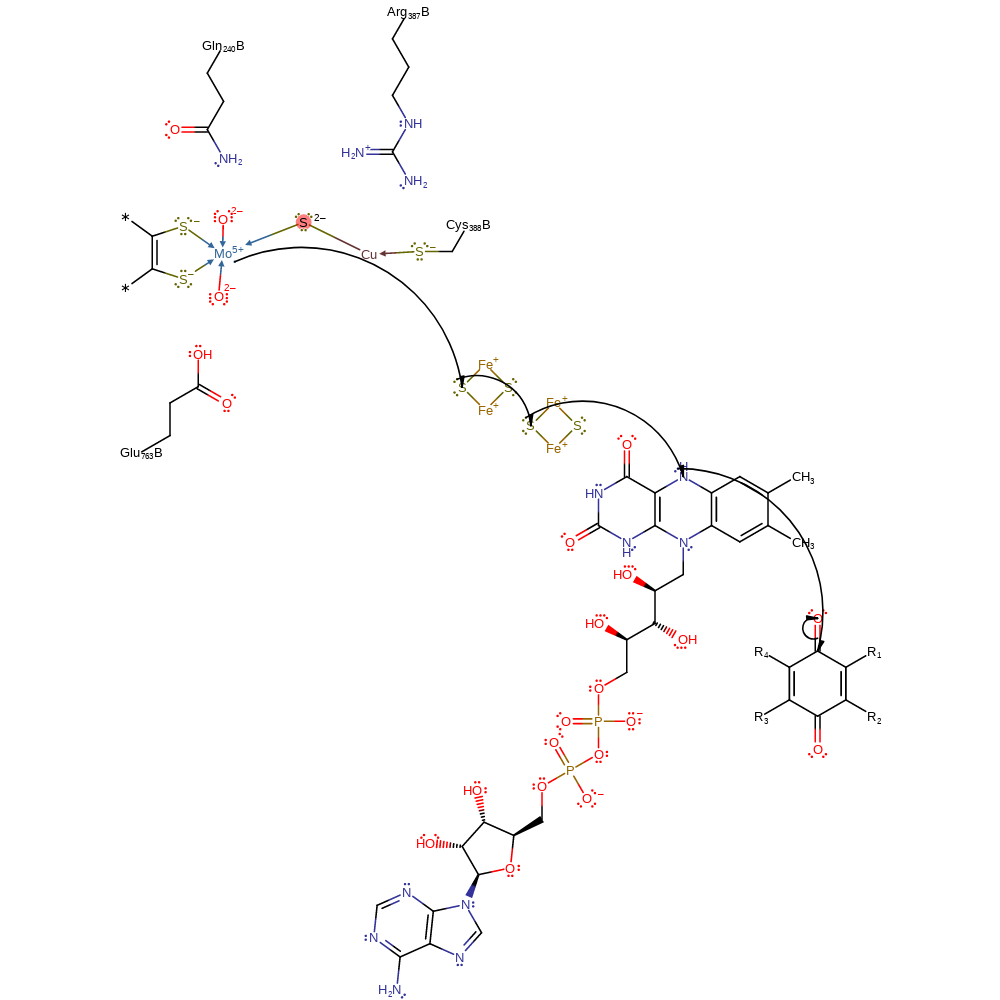
Step 6. The second single electron transfer from the Mo(V) through two iron-sulfur clusters to FAD, and thence to an external electron acceptor. This regenerates the cofactor.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg387B | electrostatic stabiliser |
| Gln240B | electrostatic stabiliser |
| Cys388B | metal ligand |
Chemical Components
electron transfer, intermediate terminated, cofactor used, native state of cofactor regenerated, overall product formed, proton relay, electron relay, inferred reaction stepCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gln240B | electrostatic stabiliser |
| Arg387B | electrostatic stabiliser |
| Cys388B | metal ligand |
| Glu763B | proton donor |
Chemical Components
inferred reaction step, native state of enzyme regenerated, proton transferIntroduction
Proposed catalytic cycle for CODH that avoids formation of a stable C-S bonded intermediate.
This mechanism starts in the oxidized Mo(VI)-Cu(I) state. The substrate carbon monoxide (CO) reaches the cluster through the substrate channel. CO coordinates to the copper (I) centre. This activates the CO and allows for the formation of the C-O bond between CO and the oxygen derived ligand on Mo(VI). This results in the reduction of the molybdenum to Mo(IV) and the formation of a five membered ring. Carbon dioxide is then released, and the molybdenum becomes a 4-coordinated Mo(IV). Reoxidation of the Mo(IV) centre by an external electron acceptor completes the proposed catalytic cycle. Electrons released are transferred via the sulfido ligand to yield the reduced Mo(IV) state. Cu(I) does not change oxidation state during the mechanism. The chain of redox active cofactors (type-I [2Fe2S] cluster, type-II [2Fe2S] cluster, and FAD) facilitates the reoxidation of the reduced Mo-ion through electron transfer from the active site Mo to external redox partners like cytochrome b561.
Catalytic Residues Roles
| UniProt | PDB* (1n62) | ||
| Glu763 | Glu763B | Glu-763 interacts through its carboxyl atom with the Mo ion in trans position to the oxo-group and is linked through hydrogen bonds to the active-site loop carrying Cys-388. This residue acts as a general acid/base in the reaction. | proton relay, proton acceptor, proton donor |
| Gln240 | Gln240B | The Neta2 group of Gln-240 is within 2.90 Angstroms hydrogen-bonding distance to the apical oxo-ligand at the Mo ion, helping to activate and stabilise the ligand. | electrostatic stabiliser |
| Cys388 | Cys388B | One of the copper ligands. Activates the copper centre. | metal ligand |
| Arg387 | Arg387B | Helps stabilise the negative charge in the active site. | electrostatic stabiliser |
Chemical Components
intramolecular nucleophilic addition, electron transfer, intermediate formation, cyclisation, proton transfer, coordination to a metal ion, bimolecular nucleophilic substitution, decarboxylation, bimolecular elimination, intermediate terminated, cofactor used, native state of cofactor regenerated, overall product formed, electron relay, inferred reaction step, proton relay, native state of enzyme regeneratedReferences
- Stein BW et al. (2014), Chem Commun (Camb), 50, 1104-1106. Orbital contributions to CO oxidation in Mo-Cu carbon monoxide dehydrogenase. DOI:10.1039/c3cc47705c. PMID:24322538.
- Pelzmann AM et al. (2014), J Biol Inorg Chem, 19, 1399-1414. Insights into the posttranslational assembly of the Mo-, S- and Cu-containing cluster in the active site of CO dehydrogenase of Oligotropha carboxidovorans. DOI:10.1007/s00775-014-1201-y. PMID:25377894.

Step 1. The Carbanion of carbon monoxide initiates a nucleophilic attack on the oxo group coordinated to the Mo(VI) centre, which results in the reduction of the molybdenum to Mo(IV). The C#O triple bond reduces in order to C=O.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg387B | electrostatic stabiliser |
| Gln240B | electrostatic stabiliser |
| Cys388B | metal ligand |
Chemical Components
ingold: intramolecular nucleophilic addition, electron transfer, intermediate formation, cyclisation
Step 2. A water molecule, activated by Glu763 ligates to the Cu(I) centre.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg387B | electrostatic stabiliser |
| Gln240B | electrostatic stabiliser |
| Cys388B | metal ligand |
| Glu763B | proton acceptor |
Chemical Components
intermediate formation, proton transfer, coordination to a metal ion
Step 3. The hydroxide anion initiates a nucleophilic attack on the carbenium ion, breaking the Cu-C coordinate bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys388B | metal ligand |
| Gln240B | electrostatic stabiliser |
| Arg387B | electrostatic stabiliser |
Chemical Components
ingold: bimolecular nucleophilic substitution
Step 4. the intermediate collapses (probably initiated by Glu763), eliminating carbon dioxide and reforming the oxo ligand on Mo(IV).
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys388B | metal ligand |
| Glu763B | proton donor, proton acceptor, proton relay |
Chemical Components
decarboxylation, ingold: bimolecular elimination, proton transfer
Step 5. A single electron transfer, through two iron-sulfur clusters to FAD, and thence to an external electron acceptor. This regenerates the cofactor.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg387B | electrostatic stabiliser |
| Gln240B | electrostatic stabiliser |
| Cys388B | metal ligand |
Chemical Components
electron transfer, intermediate terminated, cofactor used, native state of cofactor regenerated, overall product formed, electron relay, inferred reaction step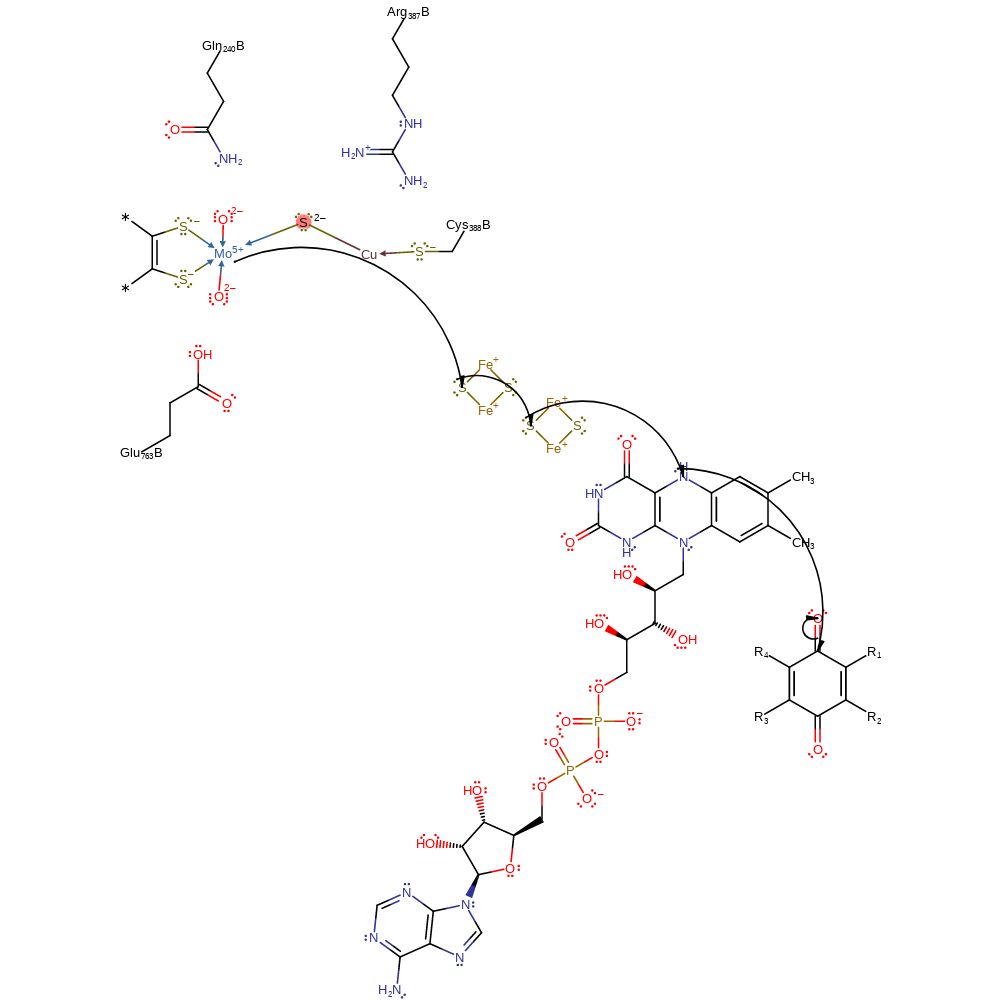
Step 6. The second single electron transfer from the Mo(V) through two iron-sulfur clusters to FAD, and thence to an external electron acceptor. This regenerates the cofactor.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gln240B | electrostatic stabiliser |
| Arg387B | electrostatic stabiliser |
| Cys388B | metal ligand |
Chemical Components
inferred reaction step, electron relay, proton relay, overall product formed, native state of cofactor regenerated, cofactor used, intermediate terminated, electron transferCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys388B | metal ligand |
| Arg387B | electrostatic stabiliser |
| Gln240B | electrostatic stabiliser |
| Glu763B | proton donor |






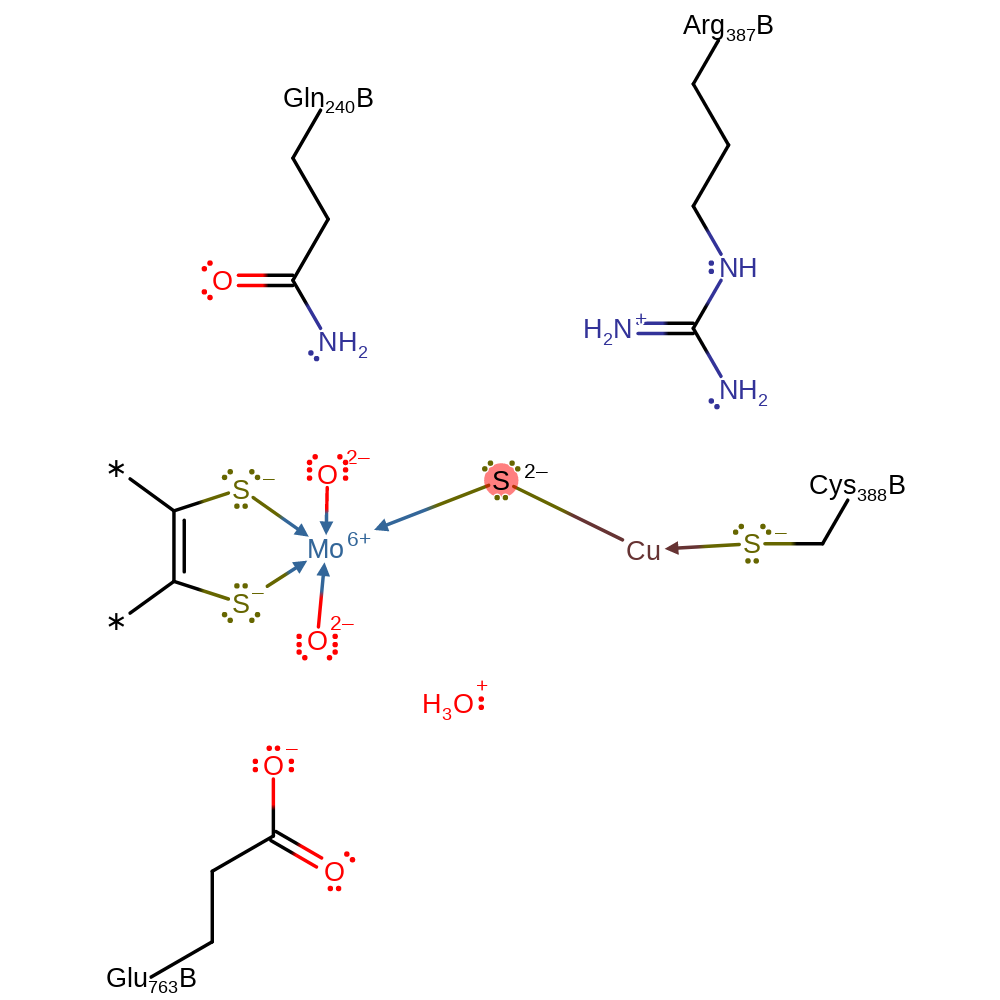 Download:
Download: 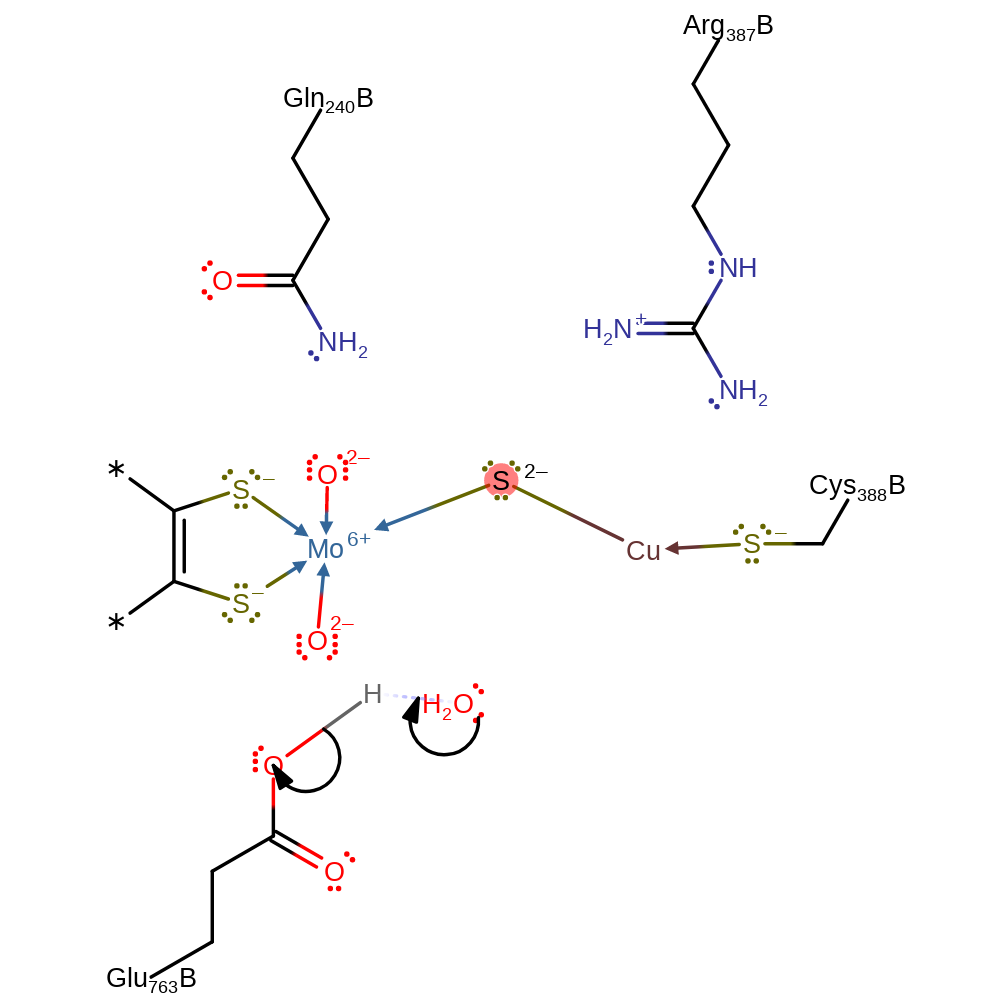
 Download:
Download: