2,4-dienoyl-CoA reductase (NADPH)
2,4-dienoyl-CoA reductase (DCR) in E. coli is an iron-sulfur flavoenzyme which contains FMN, FAD, and a 4Fe-4S cluster. It is also a monomer, unlike that of its eukaryotic counterparts which form homotetramers and lack the flavin and iron-sulfur cofactors. DCR utilises NADPH to remove the C4-C5 double bond of unsaturated fatty acids. DCR is unusual in that it lacks stereospecificity, catalysing the reduction of both natural fatty acids with cis double bonds, as well as substrates containing trans double bonds, this might be explained by the large size of the substrate binding pocket [PMID:12840019].
Reference Protein and Structure
- Sequence
-
P42593
 (1.3.1.34)
(1.3.1.34)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1ps9
- The Crystal Structure and Reaction Mechanism of E. coli 2,4-Dienoyl CoA Reductase
(2.2 Å)



- Catalytic CATH Domains
-
3.20.20.70
 (see all for 1ps9)
(see all for 1ps9)
- Cofactors
- Fadh2(2-) (1), Fmnh2(2-) (1), Tetra-mu3-sulfido-tetrairon (1) Metal MACiE
Enzyme Reaction (EC:1.3.1.34)
Enzyme Mechanism
Introduction
The reaction is initiated by hydride transfer from NADPH to FAD, which in turn transfers electrons, one at a time, to FMN via the 4Fe-4S cluster. The fully reduced FMN provides a hydride ion to the C5 atom of substrate, and Tyr and His are proposed to form a catalytic dyad that protonates the C4 atom of the substrate and completes the reaction.
Catalytic Residues Roles
| UniProt | PDB* (1ps9) | ||
| His253 | His252A | The imidazole group of His-252, which is within hydrogen bonding distance to Tyr-166 (2.7 A), is likely to stabilise the phenolate ion formed during catalysis. | hydrogen bond donor, electrostatic stabiliser |
| Arg215 | Arg214A | The reduced FMN may exist in anionic state during reduction as has been proposed, suggesting that Arg-214 would be ideally positioned to provide a counter ion. | hydrogen bond donor, electrostatic stabiliser |
| Gln340 | Gln339A | Tthe amide group of the Gln-339 side chain may participate in electron transfer between FAD and the iron-sulfur cluster by dividing electron transfer between the two cofactors from one large through-space transfer to two smaller through-space steps. However, the FAD to 4Fe-4S distance is sufficiently short that this is unlikely to be essential. | single electron relay, hydrogen bond acceptor, hydrogen bond donor, single electron acceptor, single electron donor |
| Glu165 | Glu164A | Glu-164 forms a hydrogen bond to the thioester carbonyl oxygen atom of the acyl chain, which helps to promote the positive character of the C5 atom of the acyl chain through resonance of the conjugated double bonds. | hydrogen bond donor, electrostatic stabiliser |
| Tyr167 | Tyr166A | Tyr-166 acts as the catalytic residue that protonates the C4 atom during substrate reduction. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
Chemical Components
aromatic unimolecular elimination by the conjugate base, aromatic bimolecular nucleophilic addition, hydride transfer, overall reactant used, intermediate formation, overall product formed, electron transfer, proton transfer, radical formation, cofactor used, native state of cofactor regenerated, electron relay, radical termination, bimolecular nucleophilic addition, intermediate terminated, native state of enzyme regenerated, inferred reaction stepReferences
- Hubbard PA et al. (2003), J Biol Chem, 278, 37553-37560. The Crystal Structure and Reaction Mechanism of Escherichia coli 2,4-Dienoyl-CoA Reductase. DOI:10.1074/jbc.m304642200. PMID:12840019.

Step 1. NADP eliminates a hydride ion, which is added to FAD. The nicotinamide ring of NADP interacts with the re-face of the isoalloxazine ring, resulting in fully reduced FAD [PMID:12840019].
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu164A | hydrogen bond donor |
| Tyr166A | hydrogen bond acceptor, hydrogen bond donor |
| Arg214A | hydrogen bond donor |
| His252A | hydrogen bond donor |
| Gln339A | hydrogen bond acceptor, hydrogen bond donor |
Chemical Components
ingold: aromatic unimolecular elimination by the conjugate base, ingold: aromatic bimolecular nucleophilic addition, hydride transfer, overall reactant used, intermediate formation, overall product formed
Step 2. FAD transfers a single electron, through Gln339 and an iron-sulfur cluster, to FMN, which deprotonates water.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu164A | hydrogen bond donor |
| Tyr166A | hydrogen bond acceptor, hydrogen bond donor |
| Arg214A | hydrogen bond donor |
| His252A | hydrogen bond donor |
| Gln339A | hydrogen bond acceptor, hydrogen bond donor |
| Gln339A | single electron relay, single electron donor, single electron acceptor |
Chemical Components
electron transfer, proton transfer, radical formation, overall reactant used, intermediate formation, cofactor used, native state of cofactor regenerated, electron relay
Step 3. Water deprotonates FAD, which initiates a second single electron transfer, through Gln339 and an iron-sulfur cluster, to FMN. This regenerates the FAD cofactor, and produces fully reduced FMN.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu164A | hydrogen bond donor |
| Tyr166A | hydrogen bond acceptor, hydrogen bond donor |
| Arg214A | hydrogen bond donor |
| His252A | hydrogen bond donor |
| Gln339A | hydrogen bond acceptor, hydrogen bond donor |
| Gln339A | single electron relay, single electron donor, single electron acceptor |
Chemical Components
proton transfer, electron transfer, radical termination, intermediate formation, native state of cofactor regenerated, electron relay
Step 4. FMN eliminates a hydride ion, which is added to the C5 of the substrate. This initiates double bond rearrangement, resulting in an oxyanion on the C1.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu164A | hydrogen bond donor |
| Tyr166A | hydrogen bond acceptor, hydrogen bond donor |
| Arg214A | hydrogen bond donor, electrostatic stabiliser |
| His252A | hydrogen bond donor |
| Gln339A | hydrogen bond acceptor, hydrogen bond donor |
Chemical Components
ingold: aromatic unimolecular elimination by the conjugate base, ingold: bimolecular nucleophilic addition, hydride transfer, overall reactant used, native state of cofactor regenerated, intermediate formation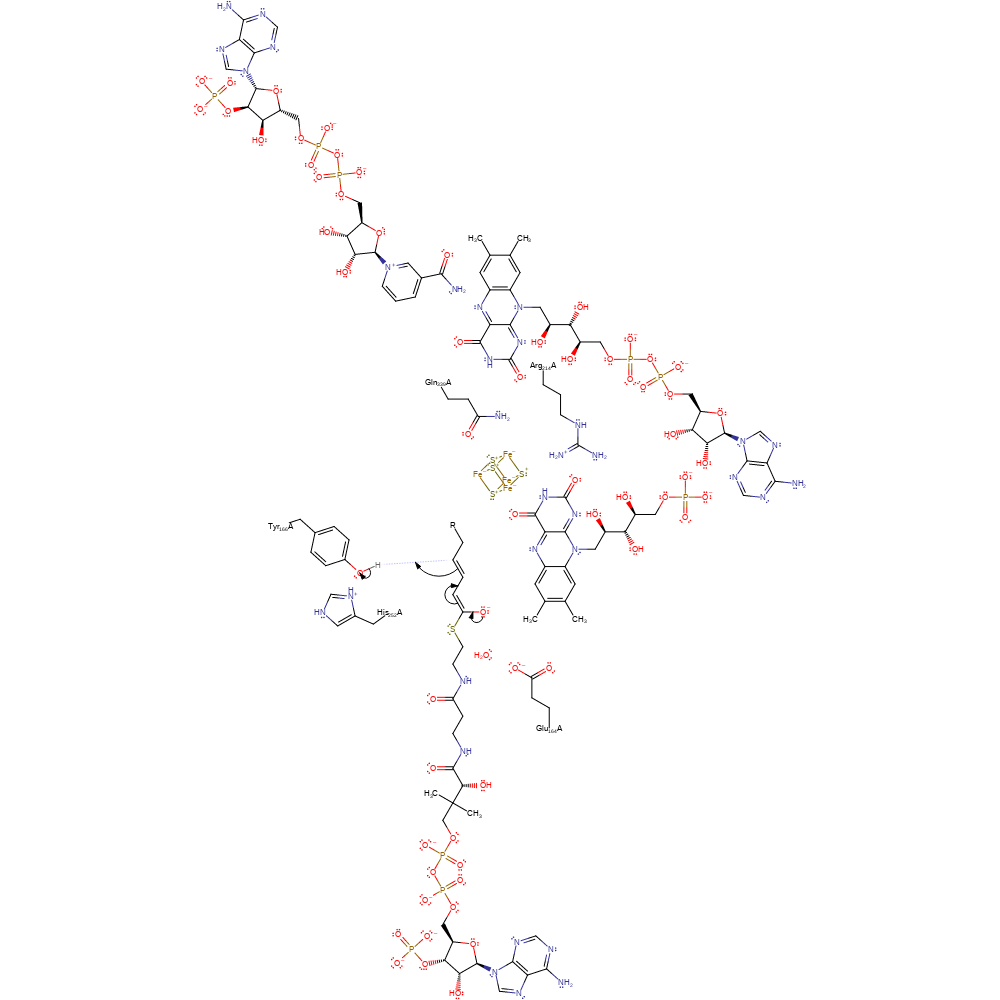
Step 5. The oxyanion collapses, initiating double bond rearrangement, the C4 of the intermediate deprotonates Tyr166.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu164A | hydrogen bond donor, electrostatic stabiliser |
| Tyr166A | hydrogen bond acceptor, hydrogen bond donor |
| Arg214A | hydrogen bond donor |
| His252A | hydrogen bond donor |
| Gln339A | hydrogen bond acceptor, hydrogen bond donor |
| Tyr166A | proton donor |
Chemical Components
proton transfer, intermediate terminated, overall product formedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr166A | hydrogen bond acceptor |
| Arg214A | hydrogen bond donor |
| His252A | hydrogen bond donor, electrostatic stabiliser |
| Gln339A | hydrogen bond acceptor, hydrogen bond donor |
| Tyr166A | proton acceptor |






 Download:
Download: