Dihydroorotate oxidase (class II)
Dihydroororate reductase in humans catalyses a key step in the synthesis of pyrimidines, as it is able to convert dihydroororate into ororate, using FMN and Ubiquinone as cofactors for the reaction. The human enzyme is part of family 2, with homology to other mammalian dihydroororate reductases and to the equivalent enzymes in bacteria which are clustered in family 1. The enzyme is particularly important in T cells because of their high nucleotide turnover through new DNA synthesis.
Reference Protein and Structure
- Sequence
-
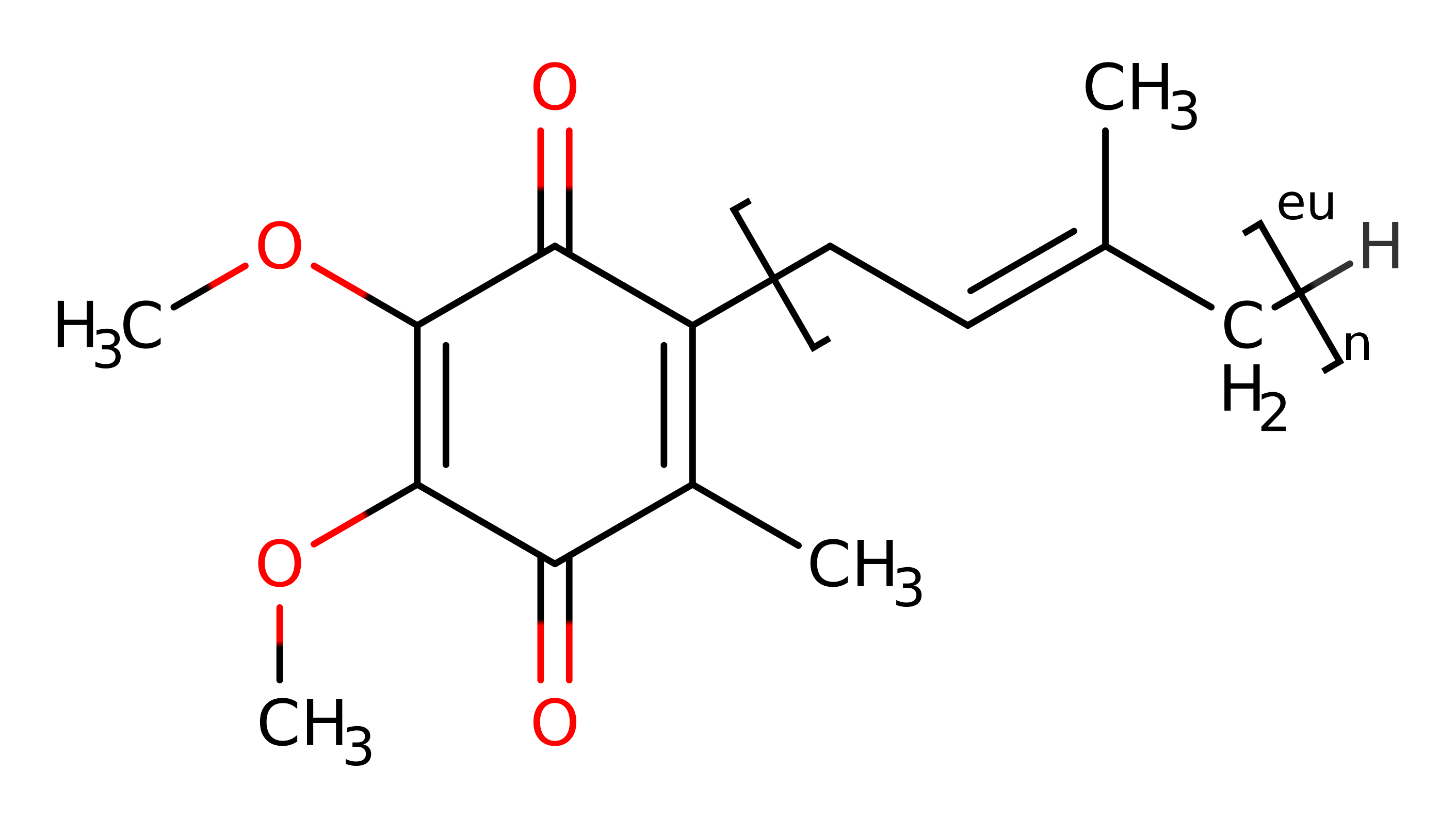
Q02127
 (1.3.5.2)
(1.3.5.2)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Homo sapiens (Human)

- PDB
-
1d3g
- HUMAN DIHYDROOROTATE DEHYDROGENASE COMPLEXED WITH BREQUINAR ANALOG
(1.6 Å)



- Catalytic CATH Domains
-
3.20.20.70
 (see all for 1d3g)
(see all for 1d3g)
- Cofactors
- Fmnh2(2-) (1)
Enzyme Reaction (EC:1.3.5.2)
Enzyme Mechanism
Introduction
In this step-wise proposal, the reaction proceeds via an enolate intermediate, which is stabilised by two highly conserved asparagines. The reaction proceeds via initial abstraction of a proton from dihydroororate by Ser 215 which is activated to act as an acid base by Thr 218 and Phe 149. This creates an oxyanion which collapses in a second step to transfers a hydride ion to FMN to form FMNH-. The hydride ion is then transferred to Ubiquinone which associates with the inner mitochondrial membrane and thus contributes to the electron transfer chain.
Catalytic Residues Roles
| UniProt | PDB* (1d3g) | ||
| Asn283, Asn216, Asn144 | Asn284(255)A, Asn217(188)A, Asn145(116)A | Help bind and stabilise the intermediates and transition states formed during the course of the reaction. | electrostatic stabiliser |
| Thr217 | Thr218(189)A | Through electrostatic contacts, is able to lower the pKa of Ser 215 to allow it to act as a general base. It has also been suggested that this residue might act as a proton relay between the bulk solvent and the active site. | activator, hydrogen bond acceptor, enhance reactivity, electrostatic stabiliser |
| Phe148 | Phe149(120)A | Through contacts between the pi electron ring and the OH group of Ser 215, is able to lower the pKa of Ser 215 sufficiently for it to act as a general base. | activator, electrostatic stabiliser, polar/non-polar interaction |
| Lys254 | Lys255(226)A | Stabilises the FMN after the transfer of a hydride ion, possibly acts as a general acid/base to allow the reduction to be completed. | hydrogen bond donor, electrostatic stabiliser |
| Ser214 | Ser215(186)A | Acts to remove proton from the dihydroororate substrate, thus allowing the carbanion to form which can transfer a hydride to the FMN cofactor. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, polar/non-polar interaction, proton relay |
Chemical Components
proton transfer, hydride transfer, assisted keto-enol tautomerisation, aromatic unimolecular elimination by the conjugate base, aromatic bimolecular nucleophilic addition, overall reactant used, native state of cofactor regenerated, intermediate terminated, overall product formed, inferred reaction step, native state of enzyme regenerated, cofactor used, native state of cofactor is not regeneratedReferences
- Fagan RL et al. (2006), Biochemistry, 45, 14926-14932. Mechanism of Flavin Reduction in Class 2 Dihydroorotate Dehydrogenases†. DOI:10.1021/bi060919g. PMID:17154530.
- Alves CN et al. (2015), Phys Chem Chem Phys, 17, 17790-17796. Insights into the mechanism of oxidation of dihydroorotate to orotate catalysed by human class 2 dihydroorotate dehydrogenase: a QM/MM free energy study. DOI:10.1039/c5cp02016f. PMID:26087682.
- Kow RL et al. (2009), Biochemistry, 48, 9801-9809. Disruption of the Proton Relay Network in the Class 2 Dihydroorotate Dehydrogenase fromEscherichia coli. DOI:10.1021/bi901024m. PMID:19694481.
- Fagan RL et al. (2009), Biochemistry, 48, 7169-7178. Roles in Binding and Chemistry for Conserved Active Site Residues in the Class 2 Dihydroorotate Dehydrogenase fromEscherichia coli. DOI:10.1021/bi900370s. PMID:19530672.
- Baumgartner R et al. (2006), J Med Chem, 49, 1239-1247. Dual Binding Mode of a Novel Series of DHODH Inhibitors. DOI:10.1021/jm0506975. PMID:16480261.

Step 1. Ser215 (activated through a triad with water and Thr218) abstracts a proton from the substrate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr218(189)A | enhance reactivity |
| Asn145(116)A | electrostatic stabiliser |
| Asn217(188)A | electrostatic stabiliser |
| Asn284(255)A | electrostatic stabiliser |
| Ser215(186)A | proton donor, proton acceptor, proton relay |
Chemical Components
proton transfer
Step 2. The enolate intermediate collapses, eliminating a hydride ion which adds onto the FMN.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn145(116)A | electrostatic stabiliser |
| Phe149(120)A | electrostatic stabiliser |
| Asn217(188)A | electrostatic stabiliser |
| Thr218(189)A | electrostatic stabiliser |
| Asn284(255)A | electrostatic stabiliser |
| Lys255(226)A | electrostatic stabiliser |
Chemical Components
hydride transfer, assisted keto-enol tautomerisation
Step 3. The product dissociates from the active site, to be replaced by ubiquinone. The reduced FMN eliminates the hydride ion which is added to the ubiquinone, initiating double bond rearrangement and deprotonation of Ser215 via a water molecue to generate ubiquinol in an inferred return step.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn284(255)A | electrostatic stabiliser |
| Asn217(188)A | electrostatic stabiliser |
| Phe149(120)A | polar/non-polar interaction, activator |
| Ser215(186)A | hydrogen bond donor, hydrogen bond acceptor, polar/non-polar interaction |
| Thr218(189)A | hydrogen bond acceptor, activator |
| Lys255(226)A | hydrogen bond donor, electrostatic stabiliser |
| Ser215(186)A | proton acceptor, proton relay, proton donor |
Chemical Components
ingold: aromatic unimolecular elimination by the conjugate base, ingold: aromatic bimolecular nucleophilic addition, hydride transfer, proton transfer, overall reactant used, native state of cofactor regenerated, intermediate terminated, overall product formed, inferred reaction step, native state of enzyme regenerated, cofactor used, native state of cofactor is not regeneratedIntroduction
In this step-wise proposal, a hydride transfer from the substrate occurs first and the reaction proceeds via an iminium intermediate. In a second step, abstraction of a proton from dihydroororate by Ser 215 which is activated to act as an acid base by Thr 218 and Phe 149 occurs to produce the final product. The hydride ion is then transferred to Ubiquinone which associates with the inner mitochondrial membrane and thus contributes to the electron transfer chain.
Catalytic Residues Roles
| UniProt | PDB* (1d3g) | ||
| Asn283, Asn216, Asn144 | Asn284(255)A, Asn217(188)A, Asn145(116)A | Help bind and stabilise the intermediates and transition states formed during the course of the reaction. | electrostatic stabiliser |
| Thr217 | Thr218(189)A | Through electrostatic contacts, is able to lower the pKa of Ser 215 to allow it to act as a general base. It has also been suggested that this residue might act as a proton relay between the bulk solvent and the active site. | activator, hydrogen bond acceptor, enhance reactivity, electrostatic stabiliser |
| Phe148 | Phe149(120)A | Through contacts between the pi electron ring and the OH group of Ser 215, is able to lower the pKa of Ser 215 sufficiently for it to act as a general base. | activator, enhance reactivity, electrostatic stabiliser, polar/non-polar interaction |
| Lys254 | Lys255(226)A | Stabilises the FMN after the transfer of a hydride ion, possibly acts as a general acid/base to allow the reduction to be completed. | hydrogen bond donor, electrostatic stabiliser |
| Ser214 | Ser215(186)A | Acts to remove proton from the dihydroororate substrate, thus allowing the carbanion to form which can transfer a hydride to the FMN cofactor. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, polar/non-polar interaction, proton donor, proton relay, electrostatic stabiliser |
Chemical Components
hydride transfer, proton transfer, assisted tautomerisation (not keto-enol), aromatic unimolecular elimination by the conjugate base, aromatic bimolecular nucleophilic addition, overall reactant used, native state of cofactor regenerated, intermediate terminated, overall product formed, inferred reaction step, native state of enzyme regenerated, cofactor used, native state of cofactor is not regeneratedReferences
- Fagan RL et al. (2006), Biochemistry, 45, 14926-14932. Mechanism of Flavin Reduction in Class 2 Dihydroorotate Dehydrogenases†. DOI:10.1021/bi060919g. PMID:17154530.
- Alves CN et al. (2015), Phys Chem Chem Phys, 17, 17790-17796. Insights into the mechanism of oxidation of dihydroorotate to orotate catalysed by human class 2 dihydroorotate dehydrogenase: a QM/MM free energy study. DOI:10.1039/c5cp02016f. PMID:26087682.
- Kow RL et al. (2009), Biochemistry, 48, 9801-9809. Disruption of the Proton Relay Network in the Class 2 Dihydroorotate Dehydrogenase fromEscherichia coli. DOI:10.1021/bi901024m. PMID:19694481.
- Fagan RL et al. (2009), Biochemistry, 48, 7169-7178. Roles in Binding and Chemistry for Conserved Active Site Residues in the Class 2 Dihydroorotate Dehydrogenase fromEscherichia coli. DOI:10.1021/bi900370s. PMID:19530672.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys255(226)A | electrostatic stabiliser |
| Phe149(120)A | enhance reactivity, polar/non-polar interaction |
| Asn145(116)A | electrostatic stabiliser |
| Asn217(188)A | electrostatic stabiliser |
| Asn284(255)A | electrostatic stabiliser |
Chemical Components
hydride transfer
Step 2. Ser215 abstracts a proton from the intermediate to form the final product.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Phe149(120)A | electrostatic stabiliser |
| Thr218(189)A | electrostatic stabiliser |
| Lys255(226)A | electrostatic stabiliser |
| Thr218(189)A | enhance reactivity |
| Asn145(116)A | electrostatic stabiliser |
| Asn217(188)A | electrostatic stabiliser |
| Asn284(255)A | electrostatic stabiliser |
| Ser215(186)A | proton acceptor, proton donor, proton relay |
Chemical Components
proton transfer, assisted tautomerisation (not keto-enol)
Step 3. The product dissociates from the active site, to be replaced by ubiquinone. The reduced FMN eliminates the hydride ion which is added to the ubiquinone, initiating double bond rearrangement and deprotonation of Ser215 via a water molecue to generate ubiquinol in an inferred return step.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Phe149(120)A | polar/non-polar interaction, activator |
| Ser215(186)A | hydrogen bond donor, hydrogen bond acceptor, polar/non-polar interaction |
| Thr218(189)A | hydrogen bond acceptor, activator |
| Lys255(226)A | hydrogen bond donor, electrostatic stabiliser |
| Ser215(186)A | electrostatic stabiliser |
| Asn284(255)A | electrostatic stabiliser |
| Ser215(186)A | proton acceptor, proton relay, proton donor |
Chemical Components
ingold: aromatic unimolecular elimination by the conjugate base, ingold: aromatic bimolecular nucleophilic addition, hydride transfer, proton transfer, overall reactant used, native state of cofactor regenerated, intermediate terminated, overall product formed, inferred reaction step, native state of enzyme regenerated, cofactor used, native state of cofactor is not regeneratedIntroduction
This mechanism proposal represents the concerted reaction. The reaction proceeds via initial abstraction of a proton from dihydroororate by Ser 215 which is activated to act as an acid base by Thr 218 and Phe 149. This creates a carbanion which transfers a hydride ion to FMN to form FMNH2, assisted by protonation of the FMN by Lys 255, forming the product. The hydride ion is then transferred to Ubiquinone which associates with the inner mitochondrial membrane and thus contributes to the electron transfer chain.
Catalytic Residues Roles
| UniProt | PDB* (1d3g) | ||
| Asn283, Asn216, Asn144 | Asn284(255)A, Asn217(188)A, Asn145(116)A | Help bind and stabilise the intermediates and transition states formed during the course of the reaction. | electrostatic stabiliser |
| Thr217 | Thr218(189)A | Through electrostatic contacts, is able to lower the pKa of Ser 215 to allow it to act as a general base. It has also been suggested that this residue might act as a proton relay between the bulk solvent and the active site. | activator, hydrogen bond acceptor |
| Phe148 | Phe149(120)A | Through contacts between the pi electron ring and the OH group of Ser 215, is able to lower the pKa of Ser 215 sufficiently for it to act as a general base. | activator, electrostatic stabiliser, polar/non-polar interaction |
| Lys254 | Lys255(226)A | Stabilises the FMN after the transfer of a hydride ion, possibly acts as a general acid/base to allow the reduction to be completed. | hydrogen bond donor, electrostatic stabiliser |
| Ser214 | Ser215(186)A | Acts to remove proton from the dihydroororate substrate, thus allowing the carbanion to form which can transfer a hydride to the FMN cofactor. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, polar/non-polar interaction, proton relay |
Chemical Components
bimolecular elimination, aromatic bimolecular nucleophilic addition, hydride transfer, overall reactant used, cofactor used, intermediate formation, overall product formed, aromatic unimolecular elimination by the conjugate base, proton transfer, native state of cofactor regenerated, intermediate terminated, inferred reaction step, native state of enzyme regenerated, native state of cofactor is not regeneratedReferences
- Liu S et al. (2000), Structure, 8, 25-33. Structures of human dihydroorotate dehydrogenase in complex with antiproliferative agents. DOI:10.1016/s0969-2126(00)00077-0. PMID:10673429.
- Kow RL et al. (2009), Biochemistry, 48, 9801-9809. Disruption of the Proton Relay Network in the Class 2 Dihydroorotate Dehydrogenase fromEscherichia coli. DOI:10.1021/bi901024m. PMID:19694481.
- Fagan RL et al. (2009), Biochemistry, 48, 7169-7178. Roles in Binding and Chemistry for Conserved Active Site Residues in the Class 2 Dihydroorotate Dehydrogenase fromEscherichia coli. DOI:10.1021/bi900370s. PMID:19530672.
- Baumgartner R et al. (2006), J Med Chem, 49, 1239-1247. Dual Binding Mode of a Novel Series of DHODH Inhibitors. DOI:10.1021/jm0506975. PMID:16480261.

Step 1. Ser215 deprotonates the C5 of dihydroorotate, eliminating a hydride ion, which is added to FMN.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Phe149(120)A | polar/non-polar interaction, electrostatic stabiliser |
| Ser215(186)A | hydrogen bond acceptor, polar/non-polar interaction |
| Thr218(189)A | hydrogen bond acceptor |
| Lys255(226)A | hydrogen bond donor |
| Asn145(116)A | electrostatic stabiliser |
| Asn217(188)A | electrostatic stabiliser |
| Asn284(255)A | electrostatic stabiliser |
| Ser215(186)A | proton acceptor, proton donor, proton relay |
Chemical Components
ingold: bimolecular elimination, ingold: aromatic bimolecular nucleophilic addition, hydride transfer, overall reactant used, cofactor used, intermediate formation, overall product formed
Step 2. The product dissociates from the active site, to be replaced by ubiquinone. The reduced FMN eliminates the hydride ion which is added to the ubiquinone, initiating double bond rearrangement and deprotonation of Ser215 via a water molecue to generate ubiquinol in an inferred return step.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Phe149(120)A | polar/non-polar interaction, activator |
| Ser215(186)A | hydrogen bond donor, hydrogen bond acceptor, polar/non-polar interaction |
| Thr218(189)A | hydrogen bond acceptor, activator |
| Lys255(226)A | hydrogen bond donor, electrostatic stabiliser |
| Asn217(188)A | electrostatic stabiliser |
| Asn284(255)A | electrostatic stabiliser |
| Ser215(186)A | proton acceptor, proton relay, proton donor |



 Download:
Download: 
 Download:
Download:  Download:
Download: