Glutamate synthase (ferredoxin)
Binds a [3Fe-4S] cluster as well as FAD and FMN. The protein is composed of two domains, one hydrolysing L-glutamine to ammonia and L-glutamate (cf. EC 3.5.1.2, glutaminase), the other combines the produced ammonia with 2-oxoglutarate to produce a second molecule of L-glutamate. The ammonia is channelled through a tunnel in the active protein. This tunnel provides a micro-environment that prevents the protonation of the ammonia, crucial as the enzyme will not function with ammonium as the nitrogen source. The hydrolysis reaction only occurs when ferredoxin and 2-oxoglutarate are bound to the protein.
Reference Protein and Structure
- Sequence
-
P55038
 (1.4.7.1)
(1.4.7.1)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Synechocystis sp. PCC 6803 substr. Kazusa (Bacteria)

- PDB
-
1ofd
- Glutamate Synthase from Synechocystis sp in complex with 2-Oxoglutarate at 2.0 Angstrom resolution
(2.0 Å)



- Catalytic CATH Domains
-
3.60.20.10
 3.20.20.70
3.20.20.70  (see all for 1ofd)
(see all for 1ofd)
- Cofactors
- Fmnh2(2-) (1), Tri-mu-sulfido-mu3-sulfido-triiron(0) (1) Metal MACiE
Enzyme Reaction (EC:1.4.7.1)
Enzyme Mechanism
Introduction
The overall transformation occurs in two halves:
- The hydrolysis of glutamine to glutamate and ammonia. This reaction occurs in the N-terminal amidotransferase domain (CATH code 3.60.20.10, residues 1-422) and proceeds via the classical cysteine covalent intermediate.
- The second half reaction occur in the FMN-binding domain (CATH code 3.20.20.70, residues 787-1223). In this reaction the ammonia initiates a nucleophilic attack on the C2 carbonyl carbon of the 2-oxoglutarate substrate. The FMN cofactor then donates a hydride ion to the intermediate and Ferredoxin regenerates the FMN cofactor.
Catalytic Residues Roles
| UniProt | PDB* (1ofd) | ||
| Cys37 (N-term) | Cys1A (N-term) | Acts as a general acid/base. It abstracts a proton (via a water relay) from its side chain in the first part of the reaction. It then donates a proton to the leaving ammonia group. In the final stages of the reaction, it abstracts a proton from a water molecule (via a water relay), finally donating the proton back to its side chain. | covalently attached, hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Asn263, Gly264 (main-N) | Asn227A, Gly228A (main-N) | Form the oxyanion hole. | hydrogen bond donor, electrostatic stabiliser |
| Glu939, Gln1005 | Glu903A, Gln969A | Activates the catalytic ammonia group. | hydrogen bond acceptor, electrostatic stabiliser |
| Lys1008 | Lys972A | Acts as a general acid/base. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, activator |
| Gln1014 (main-N) | Gln978A (main-N) | Helps stabilise the transition states formed during the course of the second half reaction. | activator, hydrogen bond donor |
| Arg67 (main-C), Phe243 (main-N) | Arg31A (main-C), Phe207A (main-N) | Help activate the water molecule that is part of the proton relay between Cys1 N-terminal group and side chain. | activator, hydrogen bond acceptor, electrostatic stabiliser |
| Cys37 | Cys1A | Acts as a general acid/base and is the catalytic nucleophile. | covalently attached, nucleofuge, nucleophile, proton acceptor, proton donor |
Chemical Components
proton transfer, bimolecular nucleophilic addition, overall reactant used, enzyme-substrate complex formation, intermediate formation, proton relay, unimolecular elimination by the conjugate base, deamination, enzyme-substrate complex cleavage, intermediate collapse, overall product formed, native state of enzyme regenerated, intermediate terminated, dehydration, schiff base formed, aromatic unimolecular elimination by the conjugate base, hydride transfer, cofactor used, inferred reaction step, redox reaction, radical formation, electron relay, electron transfer, radical termination, native state of cofactor regeneratedReferences
- van den Heuvel RH et al. (2003), J Mol Biol, 330, 113-128. The Active Conformation of Glutamate Synthase and its Binding to Ferredoxin. DOI:10.1016/s0022-2836(03)00522-9. PMID:12818206.
- Vanoni MA et al. (2008), IUBMB Life, 60, 287-300. Structure–function studies of glutamate synthases: A class of self-regulated iron-sulfur flavoenzymes essential for nitrogen assimilation. DOI:10.1002/iub.52. PMID:18421771.
- Agnelli P et al. (2005), Arch Biochem Biophys, 436, 355-366. The unexpected structural role of glutamate synthase [4Fe–4S]+1,+2 clusters as demonstrated by site-directed mutagenesis of conserved C residues at the N-terminus of the enzyme β subunit. DOI:10.1016/j.abb.2005.02.009. PMID:15797248.
- Vanoni MA et al. (2005), Arch Biochem Biophys, 433, 193-211. Structure–function studies on the iron–sulfur flavoenzyme glutamate synthase: an unexpectedly complex self-regulated enzyme. DOI:10.1016/j.abb.2004.08.033. PMID:15581577.
- van den Heuvel RH et al. (2004), Cell Mol Life Sci, 61, 669-681. Glutamate synthase: a fascinating pathway from L-glutamine to L-glutamate. DOI:10.1007/s00018-003-3316-0. PMID:15052410.
- van den Heuvel RH et al. (2002), J Biol Chem, 277, 24579-24583. Structural Studies on the Synchronization of Catalytic Centers in Glutamate Synthase. DOI:10.1074/jbc.m202541200. PMID:11967268.
- Oinonen C et al. (2000), Protein Sci, 9, 2329-2337. Structural comparison of Ntn-hydrolases. DOI:10.1110/ps.9.12.2329. PMID:11206054.

Step 1. The N-terminus of Cys1 deprotonates deprotonates water, which deprotonates the thiol group of Cys1, initiating a nucleophilic attack on the amide carbon in an addition reaction.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn227A | hydrogen bond donor, electrostatic stabiliser |
| Gly228A (main-N) | hydrogen bond donor |
| Cys1A (N-term) | hydrogen bond acceptor, hydrogen bond donor |
| Arg31A (main-C) | hydrogen bond acceptor, activator |
| Phe207A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Cys1A | proton donor |
| Cys1A (N-term) | proton acceptor |
| Cys1A | nucleophile |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, overall reactant used, enzyme-substrate complex formation, intermediate formation, proton relay
Step 2. The oxyanion initiates an elimination that cleaves ammonia from the bound L-glutamine substrate. Ammonia deprotonates water, which deprotonates the N-terminus of Cys1.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Cys1A | covalently attached |
| Asn227A | hydrogen bond donor, electrostatic stabiliser |
| Gly228A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Cys1A (N-term) | hydrogen bond donor, covalently attached, hydrogen bond acceptor |
| Arg31A (main-C) | hydrogen bond acceptor, electrostatic stabiliser |
| Phe207A (main-N) | hydrogen bond donor |
| Cys1A (N-term) | proton donor |
Chemical Components
proton transfer, ingold: unimolecular elimination by the conjugate base, proton relay, deamination, enzyme-substrate complex cleavage, intermediate collapse, intermediate formation
Step 3. The N-terminus of Cys1 deprotonates water, which deprotonates another water that initiates a nucleophilic attack on the carbonyl carbon of the covalently bound intermediate in an addition reaction.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn227A | hydrogen bond donor, electrostatic stabiliser |
| Gly228A (main-N) | hydrogen bond donor |
| Cys1A (N-term) | hydrogen bond acceptor, covalently attached |
| Arg31A (main-C) | hydrogen bond acceptor, activator |
| Phe207A (main-N) | hydrogen bond donor |
| Cys1A (N-term) | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, proton relay, enzyme-substrate complex formation, intermediate formation
Step 4. The oxyanion initiates an elimination that cleaves the C-S bond, the thiolate of Cys1 deprotonates water, which deprotonates the N-terminus of Cys1
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn227A | hydrogen bond donor, electrostatic stabiliser |
| Gly228A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Cys1A (N-term) | hydrogen bond donor |
| Arg31A (main-C) | hydrogen bond acceptor, electrostatic stabiliser |
| Phe207A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Cys1A (N-term) | proton donor |
| Cys1A | nucleofuge, proton acceptor |
Chemical Components
proton transfer, ingold: unimolecular elimination by the conjugate base, overall product formed, native state of enzyme regenerated, enzyme-substrate complex cleavage, proton relay, intermediate terminated, intermediate collapse
Step 5. Ammonia initiates a nucleophilic attack on the C2 carbonyl carbon of the 2-oxoglutarate substrate in an addition reaction. The oxyanion formed deprotonates the bound ammonium.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gln969A | hydrogen bond acceptor |
| Gln978A (main-N) | hydrogen bond donor, activator |
| Lys972A | hydrogen bond donor, activator |
| Glu903A | hydrogen bond acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, overall reactant used, intermediate formation
Step 6. The amine initiates an elimination of the bound hydroxide as water, which deprotonates Lys972.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gln969A | hydrogen bond acceptor |
| Gln978A (main-N) | hydrogen bond donor, activator |
| Lys972A | hydrogen bond donor |
| Glu903A | hydrogen bond acceptor |
| Lys972A | proton donor |
Chemical Components
ingold: unimolecular elimination by the conjugate base, proton transfer, intermediate collapse, intermediate formation, dehydration, schiff base formed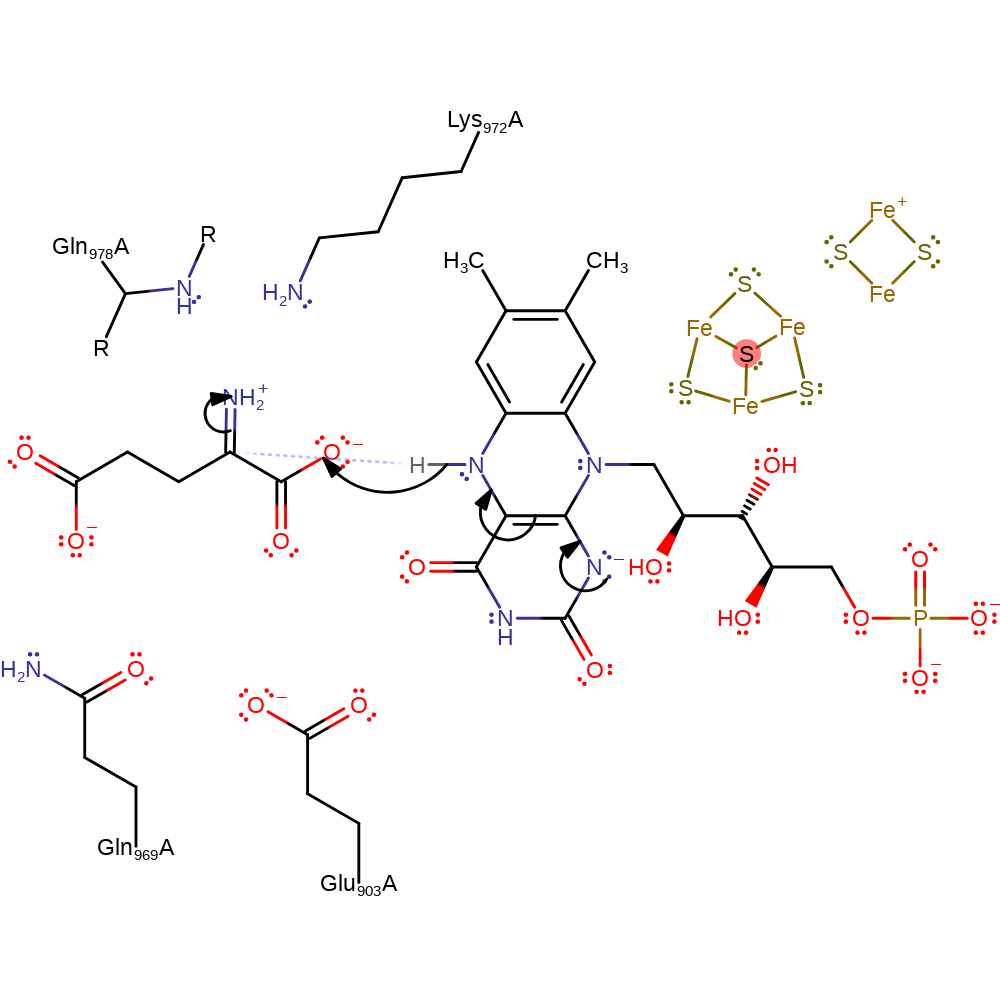
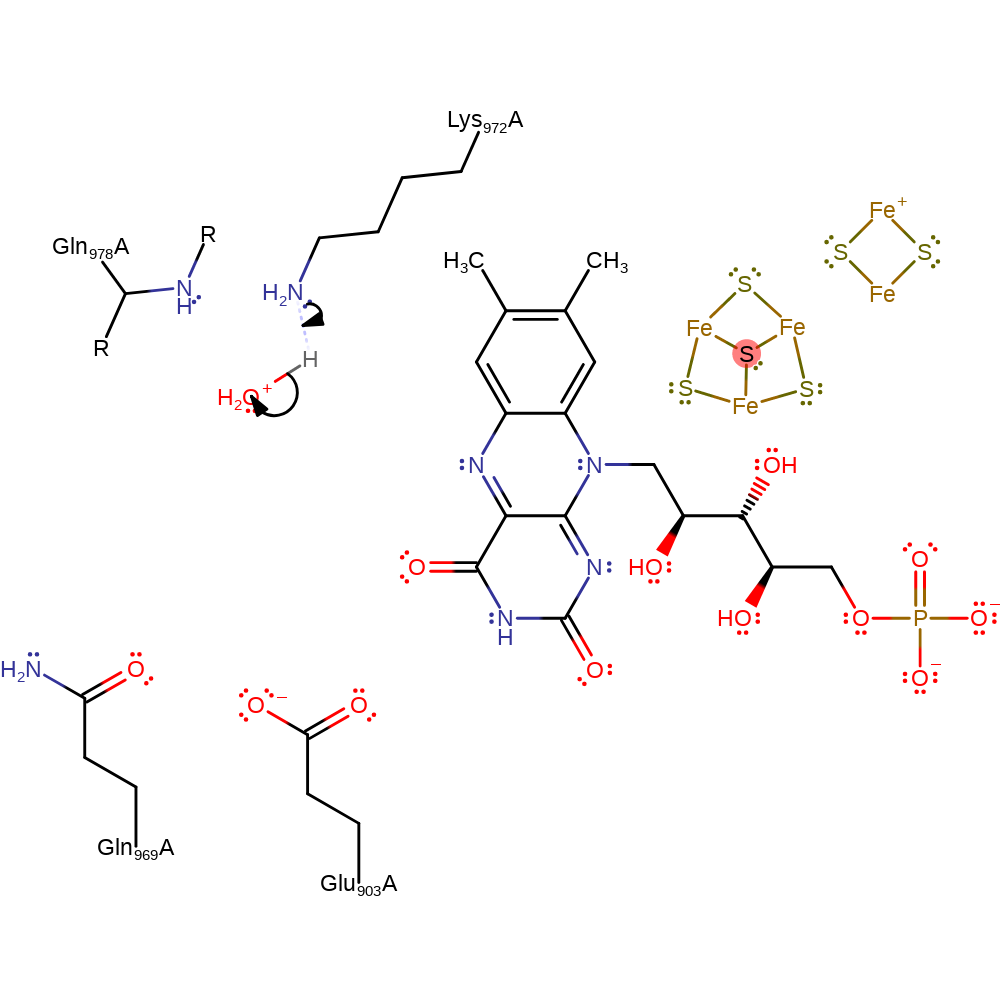
Step 7. FMN eliminates a hydride ion, which adds to the C2 imine carbon in an addition reaction.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gln969A | hydrogen bond acceptor, electrostatic stabiliser |
| Lys972A | hydrogen bond donor |
| Glu903A | hydrogen bond acceptor, electrostatic stabiliser |
Chemical Components
ingold: aromatic unimolecular elimination by the conjugate base, hydride transfer, ingold: bimolecular nucleophilic addition, cofactor used, intermediate terminated, overall product formedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys972A | hydrogen bond acceptor, proton acceptor |
Chemical Components
proton transfer, inferred reaction step
Step 9. Ferredoxin donates a single electron, which is transferred to FMN through an iron-sulfur cluster. FMN deprotonates water.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
proton transfer, redox reaction, radical formation, overall reactant used, intermediate formation, overall product formed, electron relay
Step 10. A second ferredoxin donates a single electron, which is transferred to FMN through an iron-sulfur cluster, which regenerates the reduced form of FMN.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|


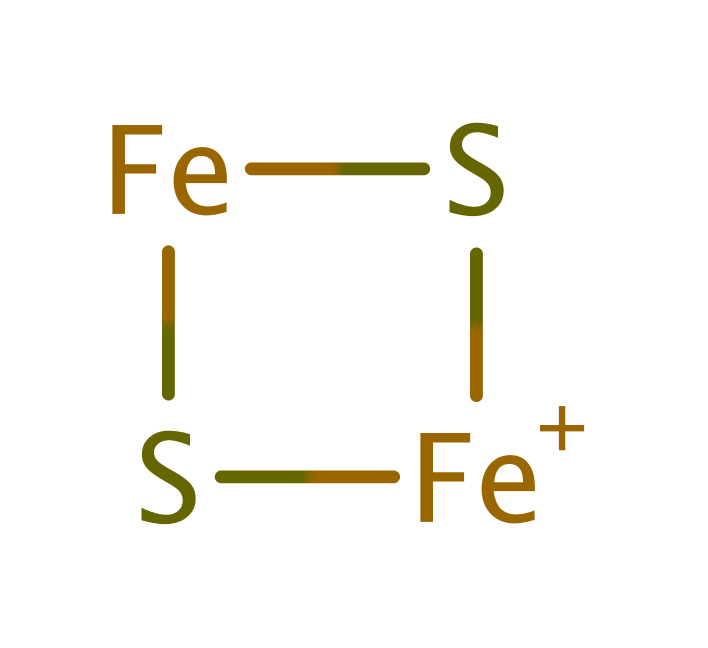


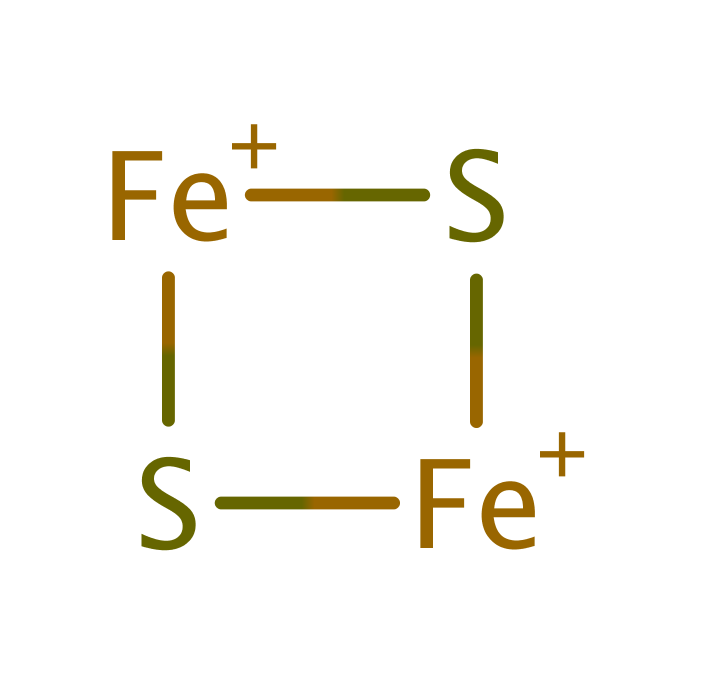
 Download:
Download: