Naphthalene 1,2-dioxygenase
The naphthalene dioxygenase (NDO) multicomponent enzyme system catalyses the incorporation of both atoms of molecular oxygen into naphthalene to form cis-naphthalene dihydrodiol. The enzyme comprises an NADH binding reductase, a Rieske feredoxin, and a oxygenase component. All three units contain [2Fe-2S] clusters. NDO is a member of the ring-hydroxylating dioxygenase (RHD) family of bacterial enzymes that play a critical role in the degradation of aromatic compounds.
Reference Protein and Structure
- Sequences
-
P0A110
 (1.14.12.12)
(1.14.12.12)
P0A112
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Pseudomonas putida (Bacteria)

- PDB
-
1ndo
- NAPHTHALENE 1,2-DIOXYGENASE
(2.25 Å)



- Catalytic CATH Domains
-
2.102.10.10
 3.90.380.10
3.90.380.10  (see all for 1ndo)
(see all for 1ndo)
- Cofactors
- Iron(3+) (1), Di-mu-sulfido-diiron(2+) (3), Fadh2(2-) (1) Metal MACiE
Enzyme Reaction (EC:1.14.12.12)
Enzyme Mechanism
Introduction
NAD initiates a hydride transfer to the FAD cofactor, which then undergoes a rearrangement to transfer a single electron to the final of three iron-sulfur clusters. This electron is then transferred to a dixygen molecule via a three residue relay (His104, Asp205 and His208). The peroxo group formed then attacks the naphthalene substrate in a nucleophilic addition that causes the double bond to attack the peroxo group, cleaving the O-O bond and forming the epoxide intermediate. This intermediate then collapses in a heterolysis resulting in a carbocation and anioinc oxygen bound to the Fe(III) centre. The iron-bound hydroxide group attacks the carbocation in a coordination reaction. The second electron transfer is initiated by the deprotonation of the FAD and occurs in the same manner as the first. In the final step of the reaction the cis-1,2-dihydronaphthalene-1,2-diol product deprotonates a water molecule and de-coordinates from the Fe(II) centre.
Catalytic Residues Roles
| UniProt | PDB* (1ndo) | ||
| His104, Asp205 | His104A, Asp205E | Forms part of the electron relay chain between the final iron sulfur cluster and the active site iron and dioxygen moleucles. | single electron relay, single electron acceptor, single electron donor |
| His208 | His208E | Forms part of the catalytic iron binding site and acts as an electron relay from the final iron sulfur cluster to the dioxygen molecule. | single electron relay, single electron acceptor, single electron donor, metal ligand |
| His213, Asp362 | His213E, Asp362E | Forms part of the catalytic iron binding site. | metal ligand |
Chemical Components
aromatic unimolecular elimination by the conjugate base, aromatic bimolecular nucleophilic addition, hydride transfer, overall reactant used, cofactor used, intermediate formation, overall product formed, redox reaction, radical formation, electron relay, electron transfer, proton transfer, bimolecular nucleophilic addition, coordination, native state of cofactor regenerated, coordination to a metal ion, bimolecular nucleophilic substitution, rate-determining step, heterolysis, decoordination from a metal ion, intermediate terminated, native state of enzyme regeneratedReferences
- Karlsson A et al. (2003), Science, 299, 1039-1042. Crystal Structure of Naphthalene Dioxygenase: Side-on Binding of Dioxygen to Iron. DOI:10.1126/science.1078020. PMID:12586937.
- Dawson WK et al. (2016), PLoS One, 11, e0162031-. Electron Transport in a Dioxygenase-Ferredoxin Complex: Long Range Charge Coupling between the Rieske and Non-Heme Iron Center. DOI:10.1371/journal.pone.0162031. PMID:27656882.
- White MD et al. (2016), Curr Opin Chem Biol, 31, 126-135. Catalytic strategies of the non-heme iron dependent oxygenases and their roles in plant biology. DOI:10.1016/j.cbpa.2016.02.017. PMID:27015291.
- Barry SM et al. (2013), ACS Catal, 3,Mechanism and Catalytic Diversity of Rieske Non-Heme Iron-Dependent Oxygenases. DOI:10.1021/cs400087p. PMID:24244885.
- Ashikawa Y et al. (2012), BMC Struct Biol, 12, 15-. Structural insight into the substrate- and dioxygen-binding manner in the catalytic cycle of rieske nonheme iron oxygenase system, carbazole 1,9a-dioxygenase. DOI:10.1186/1472-6807-12-15. PMID:22727022.
- Bassan A et al. (2004), J Biol Inorg Chem, 9, 439-452. A theoretical study of the cis -dihydroxylation mechanism in naphthalene 1,2-dioxygenase. DOI:10.1007/s00775-004-0537-0. PMID:15042436.
- Wolfe MD et al. (2003), J Biol Chem, 278, 829-835. Hydrogen Peroxide-coupled cis-Diol Formation Catalyzed by Naphthalene 1,2-Dioxygenase. DOI:10.1074/jbc.m209604200. PMID:12403773.
- Carredano E et al. (2000), J Mol Biol, 296, 701-712. Substrate binding site of naphthalene 1,2-dioxygenase: functional implications of indole binding. DOI:10.1006/jmbi.1999.3462. PMID:10669618.
- Parales RE et al. (1999), J Bacteriol, 181, 1831-1837. Aspartate 205 in the catalytic domain of naphthalene dioxygenase is essential for activity. PMID:10074076.
- Kauppi B et al. (1998), Structure, 6, 571-586. Structure of an aromatic-ring-hydroxylating dioxygenase – naphthalene 1,2-dioxygenase. DOI:10.1016/s0969-2126(98)00059-8. PMID:9634695.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp362E | metal ligand |
| His208E | metal ligand |
| His213E | metal ligand |
Chemical Components
ingold: aromatic unimolecular elimination by the conjugate base, ingold: aromatic bimolecular nucleophilic addition, hydride transfer, overall reactant used, cofactor used, intermediate formation, overall product formed
Step 2. FAD undergoes a double bond rearrangement that results in the transfer of a single electron from the FAD to one of the iron-sulfur clusters. This then initiates an electron relay through a second iron-sulfur cluster to a third.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp362E | metal ligand |
| His208E | metal ligand |
| His213E | metal ligand |
Chemical Components
redox reaction, radical formation, cofactor used, intermediate formation, electron relay
Step 3. The electron is transferred from the final iron-sulfur cluster to a dioxygen molecule, which also accepts a second electron from another Fe(II) centre, and deprotonates a water molecule.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp205E | single electron donor |
| Asp362E | metal ligand |
| His208E | metal ligand |
| His213E | metal ligand |
| His104A | single electron relay |
| Asp205E | single electron relay |
| His208E | single electron relay, single electron acceptor, single electron donor |
| His104A | single electron donor, single electron acceptor |
| Asp205E | single electron acceptor |
Chemical Components
electron transfer, proton transfer, ingold: bimolecular nucleophilic addition, coordination, overall reactant used, native state of cofactor regenerated, coordination to a metal ion, intermediate formation, electron relay
Step 4. The peroxo group attacks the naphthalene substrate in a nucleophilic addition that causes the double bond to attack the peroxo group, cleaving the O-O bond and forming the epoxide intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp362E | metal ligand |
| His208E | metal ligand |
| His213E | metal ligand |
Chemical Components
ingold: bimolecular nucleophilic addition, ingold: bimolecular nucleophilic substitution, overall reactant used, intermediate formation, rate-determining step
Step 5. The epoxide intermediate collapses in a heterolysis resulting in a carbocation and anioinc oxygen bound to the Fe(III) centre.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp362E | metal ligand |
| His208E | metal ligand |
| His213E | metal ligand |
Chemical Components
heterolysis, intermediate formation
Step 6. The iron-bound hydroxide group attacks the carbocation in a coordination reaction.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp362E | metal ligand |
| His208E | metal ligand |
| His213E | metal ligand |
Chemical Components
coordination, intermediate formation
Step 7. An unidentified base deprotonates the FAD, initiating a single electron transfer to the one of the iron-sulfur clusters. This then initiates an electron relay through a second iron-sulfur cluster to a third.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp362E | metal ligand |
| His208E | metal ligand |
| His213E | metal ligand |
Chemical Components
proton transfer, electron transfer, native state of cofactor regenerated, cofactor used, intermediate formation, electron relay
Step 8. The electron is transferred from the final iron-sulfur cluster to the Fe(III) centre
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp362E | metal ligand |
| His208E | metal ligand |
| His213E | metal ligand |
Chemical Components
electron transfer, native state of cofactor regenerated, electron relay, intermediate formation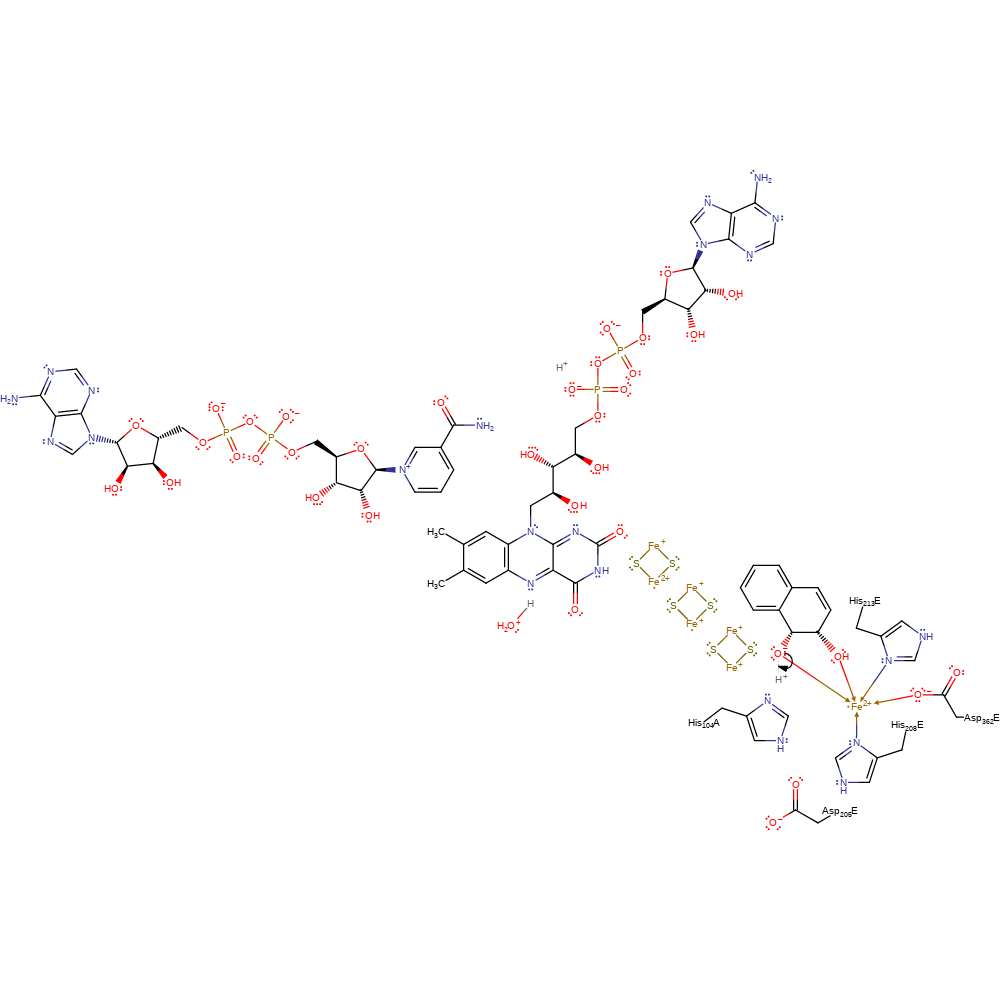
Step 9. The cis-1,2-dihydronaphthalene-1,2-diol product deprotonates a water molecule and de-coordinates from the Fe(II) centre.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp362E | metal ligand |
| His208E | metal ligand |
| His213E | metal ligand |






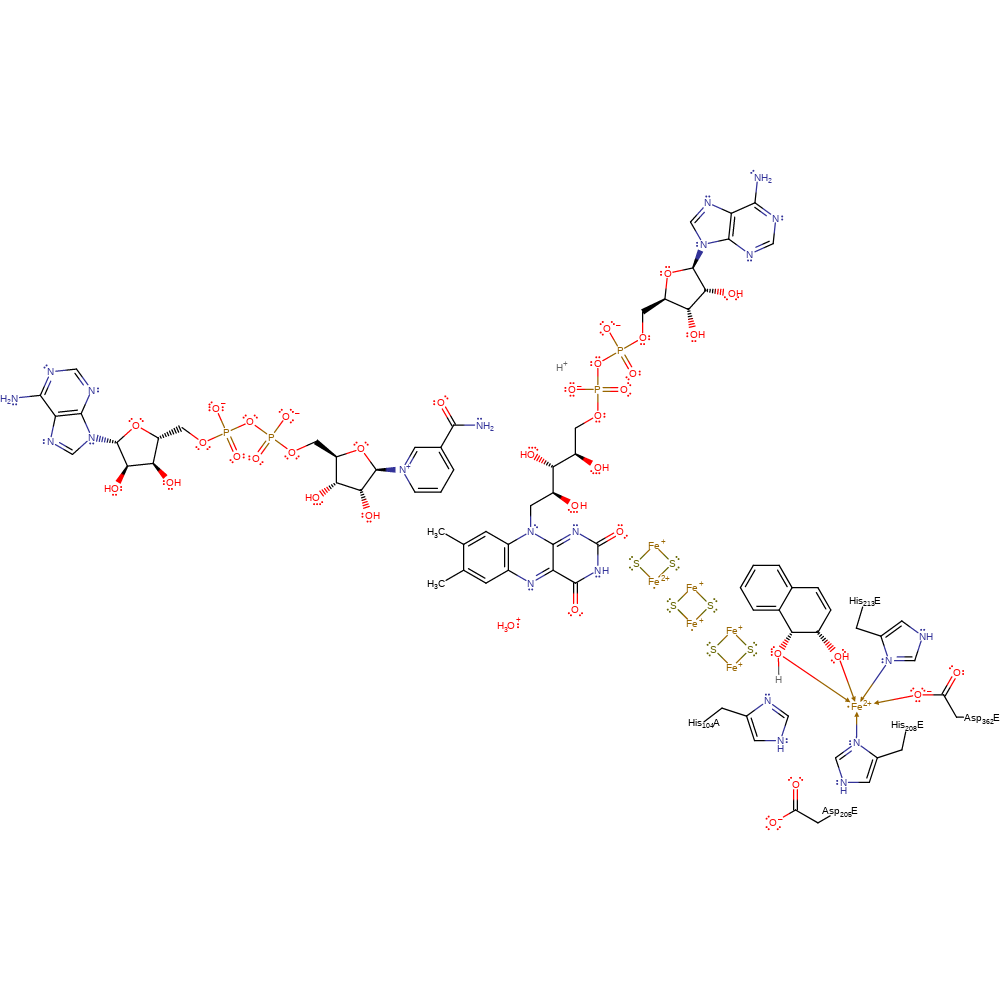 Download:
Download: