Pyrogallol hydroxytransferase
The Mo enzyme pyrogallol-phloroglucinol transhydroxylase (TH) from the anaerobic microorganism Pelobacter acidigallici catalyses the conversion of pyrogallol to phloroglucinol. Such trihydroxybenzenes and their derivatives represent important building blocks of plant polymers.
The overall reaction appears as transfer of the 2-hydroxy group of 1,2,3,5-tetrahydroxybenzene to the C5 position of pyrogallol (systematic name 1,2,3-trihydroxybenzene). This converts pyrogallol into 1,2,3,5-tetrahydroxybenzene, and 1,2,3,5-tetrahydroxybenzene into phloroglucinol (systematic name 1,3,5-trihydroxybenzene). Therefore, the net reaction consumes no 1,2,3,5-tetrahydroxybenzene.
TH is cytoplasmic and consists of a large alpha subunit and a small beta subunit. The beta subunit contains several Fe-S clusters, which have no known function and may be evolutionary relics. This subunit also has a fibronectin-like domain which can be used for membrane association. The alpha subunit contain the Mo-binding site (with Mo coordinated to two molybdopterin guanine dinucleotide (MGD) cofactors) and the catalytic active site. Like other members of the DMSO-reducing enzyme family, Mo cycles between +4 and +6 oxidation states for catalytic activity.
Reference Protein and Structure
- Sequence
-
P80563
 (1.97.1.2)
(1.97.1.2)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Pelobacter acidigallici (Bacteria)

- PDB
-
4v4e
- Crystal Structure of Pyrogallol-Phloroglucinol Transhydroxylase from Pelobacter acidigallici complexed with inhibitor 1,2,4,5-tetrahydroxy-benzene
(2.0 Å)



- Catalytic CATH Domains
- (see all for 4v4e)
- Cofactors
- Molybdenum(4+) (1), Molybdenum(6+) (1), Molybdopterin guanine dinucleotide (1) Metal MACiE
Enzyme Reaction (EC:1.97.1.2)
Enzyme Mechanism
Introduction
His 144 deprotonates pyrogallol at the O1-hydroxy group. The deprotonated oxygen atom bonds to Mo(VI). Mo(VI) oxidises the enol tautomer of pyrogallol to the orthoquinone form, getting reduced to Mo(IV). The C1 proton is transferred to Asp 174. Tyr 404 deprotonates O2 of 1,2,3,5-tetrahydroxybenzene. The deprotonated O2 attacks C5 of pyrogallol in a nucleophilic manner. This creates a diphenylether bridging bond between the two substrates; the pyrogallol C1 carbonyl deprotonates His 144 to regenerate the C1 hydroxy group. The C5 pyrogallol proton is removed (base not specified, possibly solvent) leading to regeneration of pyrogallol aromaticity and an enolate negative charge on the C2 oxygen of pyrogallol. The C5 hydroxy group of tetrahydroxybenzene is deprotonated (base not specified), converting it to the carbonyl and leading to C2 protonation and loss of aromaticity. The pyrogallol enolate collapses, leading to cleavage of the bridging phenylether bond. The bridging oxygen atom remains on pyrogallol (now a tetrahydroxybenzene quinone) as a carbonyl. Protonation of O5 of the (former) tetrahydroxybenzene means that it is now fully converted to phloroglucinol and is released. The pyrogallol quinone is reduced by Mo(IV) to the product, 1,2,3,5-tetrahydroxybenzene. These steps are not outlined in the literature. O5 is inferred to be protonated by Tyr 404, and O2 by Asp 174.
Catalytic Residues Roles
| UniProt | PDB* (4v4e) | ||
| His144 | His144A | His 244 acts as a general base and acid to the 1-hydroxyl group of pyrogallol. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, electrostatic stabiliser |
| Tyr404 | Tyr404A | Tyr 404 acts as a general base and acid to the transferred hydroxy group. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, electrostatic stabiliser |
| Asp174 | Asp174A | Asp 174 acts as a general base and acid to the 2-hydroxyl group of pyrogallol. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, electrostatic stabiliser |
Chemical Components
proton transfer, bimolecular nucleophilic substitution, overall reactant used, cofactor used, coordination to a metal ion, decoordination from a metal ion, intermediate formation, electron transfer, bimolecular nucleophilic addition, assisted keto-enol tautomerisation, intermediate terminated, unimolecular elimination by the conjugate base, intermediate collapse, overall product formed, redox reaction, inferred reaction step, radical termination, native state of cofactor regenerated, native state of enzyme regeneratedReferences
- Messerschmidt A et al. (2004), Proc Natl Acad Sci U S A, 101, 11571-11576. Crystal structure of pyrogallol-phloroglucinol transhydroxylase, an Mo enzyme capable of intermolecular hydroxyl transfer between phenols. DOI:10.1073/pnas.0404378101. PMID:15284442.
- Paizs C et al. (2007), Chemistry, 13, 2805-2811. Investigation of the mechanism of action of pyrogallol-phloroglucinol transhydroxylase by using putative intermediates. DOI:10.1002/chem.200601053. PMID:17201004.
- Hille R et al. (1998), FEMS Microbiol Rev, 22, 489-501. Mechanistic aspects of molybdenum-containing enzymes. DOI:10.1016/s0168-6445(98)00037-0. PMID:10189201.

Step 1. His144 deprotonates the pyrogallol substrate at the C3-OH position, the activated substrate then attacks the molybdenum cofactor in a nucleophilic substitution, resulting in the loss of a water molecule from the Mo(VI) coordination sphere.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His144A | hydrogen bond acceptor |
| Asp174A | hydrogen bond acceptor, electrostatic stabiliser |
| Tyr404A | hydrogen bond acceptor |
| His144A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic substitution, overall reactant used, cofactor used, coordination to a metal ion, decoordination from a metal ion, intermediate formation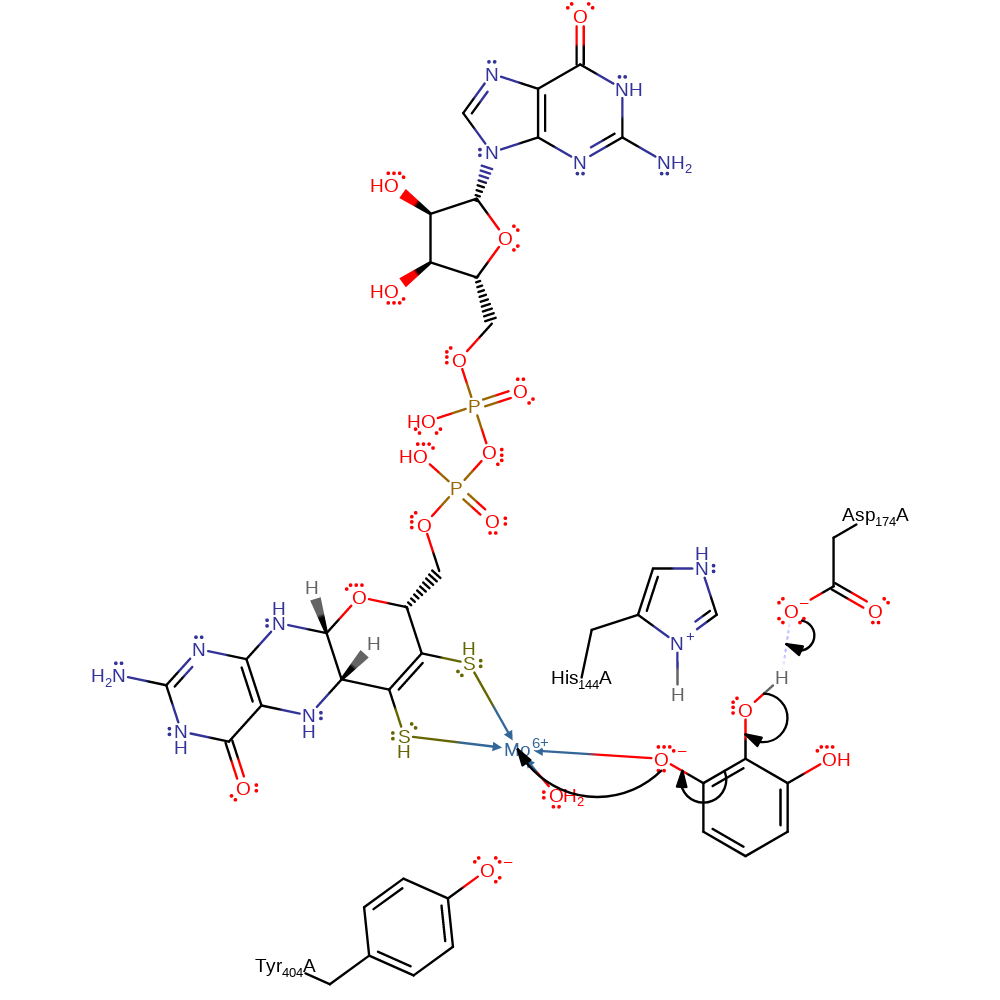
Step 2. Asp174 deprotonates the bound intermediate at the C2-OH position, resulting in a isomerisation to form the diquinone form of the substrate, and a two electron reduction of the molybdenum cofactor resulting in an Mo(IV) oxidation state.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His144A | hydrogen bond donor, electrostatic stabiliser |
| Asp174A | hydrogen bond acceptor |
| Tyr404A | hydrogen bond acceptor |
| Asp174A | proton acceptor |
Chemical Components
proton transfer, electron transfer, intermediate formation
Step 3. Tyr404 deprotonates the second substrate (1,2,3,5-tetrahydroxybenzene) at the C2-OH position.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His144A | hydrogen bond donor |
| Asp174A | hydrogen bond donor |
| Tyr404A | hydrogen bond acceptor |
| Tyr404A | proton acceptor |
Chemical Components
proton transfer, overall reactant used, intermediate formation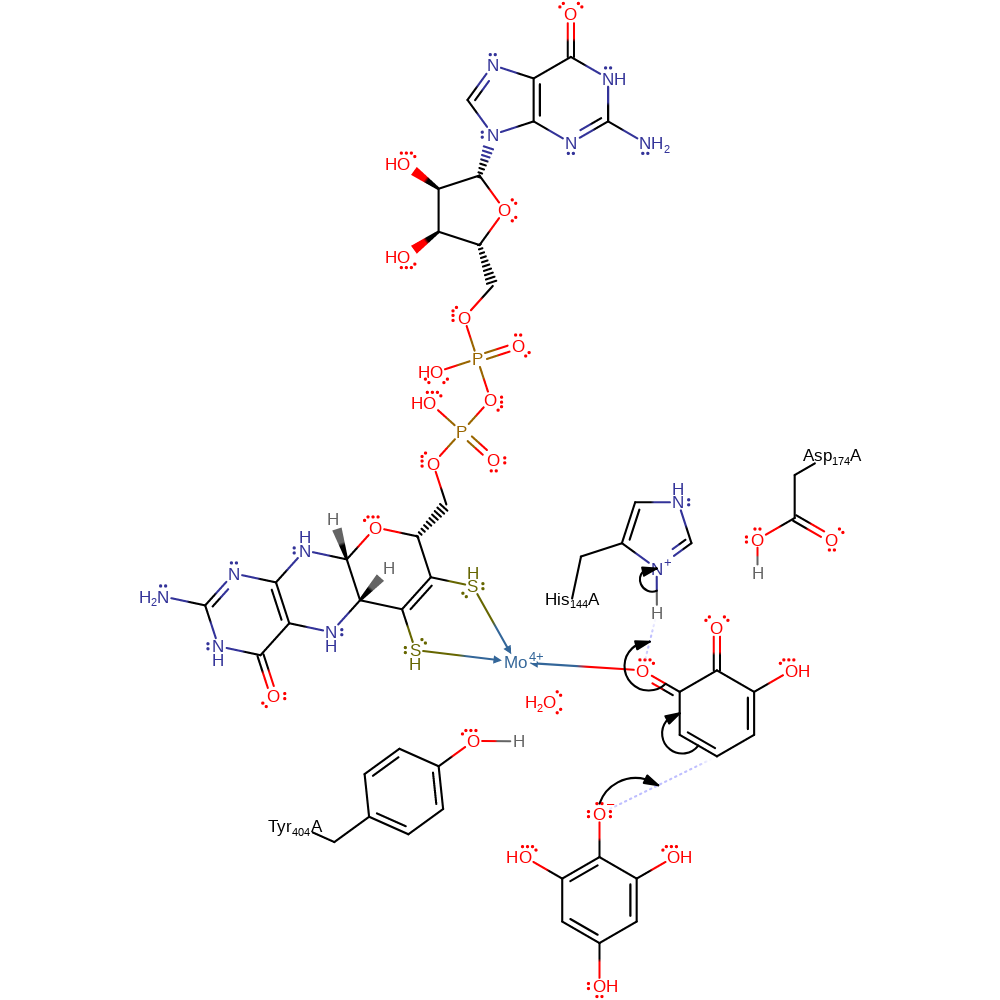
Step 4. The activated 1,2,3,5-tetrahydroxybenzene then attacks the Mo-coordinated intermediate in a nucleophilic addition, which is reprotonated from His144 at the C2=O position.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His144A | hydrogen bond donor |
| Asp174A | hydrogen bond donor |
| Tyr404A | hydrogen bond donor, electrostatic stabiliser |
| His144A | proton donor |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer, intermediate formation
Step 5. Water deprotonates the C5 position of the original substrate, resulting in double bond isomerisation.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His144A | hydrogen bond acceptor |
| Asp174A | hydrogen bond donor, electrostatic stabiliser |
| Tyr404A | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
assisted keto-enol tautomerisation, proton transfer, intermediate formation
Step 6. Water deprotonates the C5 hydroxyl of the original 1,2,3,5-tetrahydroxybenzene molecule, resulting in double bond isomerisations that lead to the protonation of the C2 position of the original 1,2,3,5-tetrahydroxybenzene molecule from water.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His144A | hydrogen bond acceptor |
| Asp174A | hydrogen bond donor, electrostatic stabiliser |
| Tyr404A | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
proton transfer, assisted keto-enol tautomerisation, intermediate terminated, intermediate formation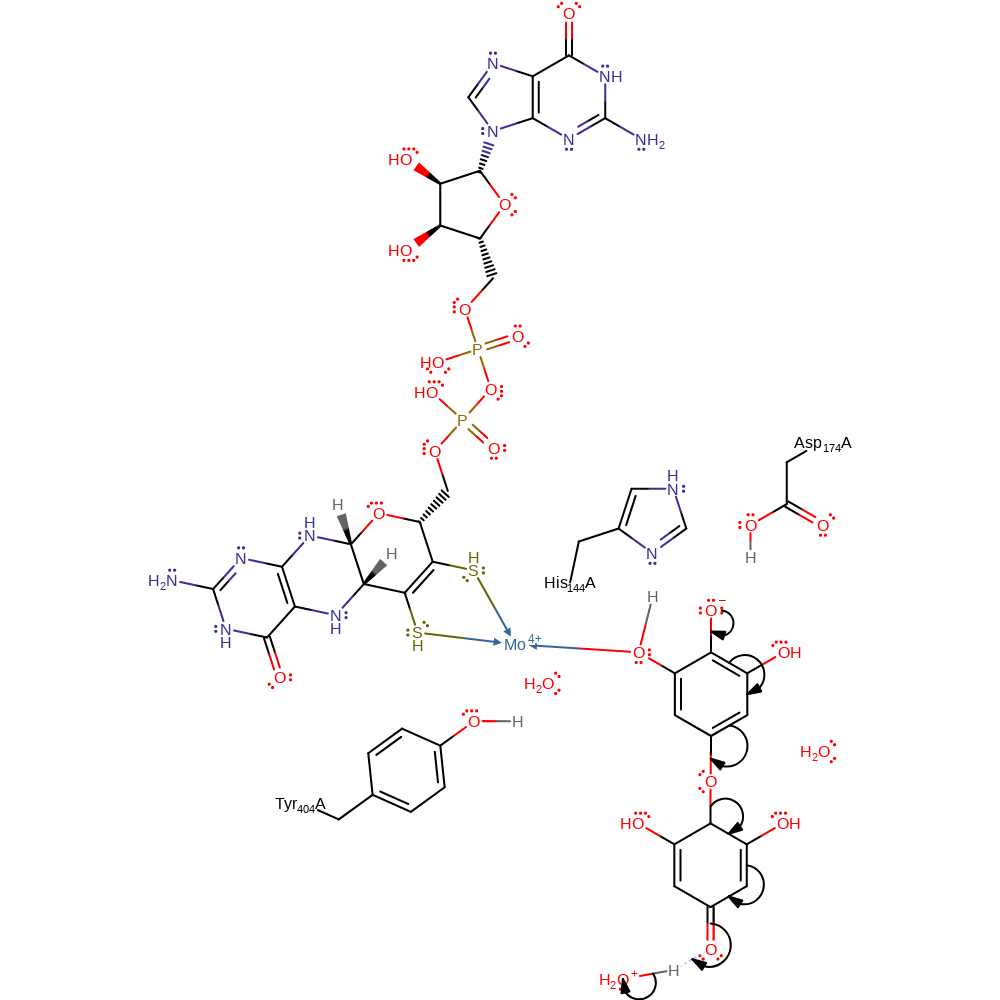
Step 7. The negatively charged oxygen at the 2 position of the original pyrogallol substrate initiates an elimination reaction which produces the diquinone form of 1,2,3,5-tetrahydroxybenzene (this is the molecule that was pyrogallol) and the product phloroglucinol from the molecule that was 1,2,3,5-tetrahydroxybenzene
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His144A | hydrogen bond acceptor |
| Asp174A | hydrogen bond donor, electrostatic stabiliser |
| Tyr404A | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
ingold: unimolecular elimination by the conjugate base, proton transfer, intermediate collapse, intermediate formation, intermediate terminated, overall product formed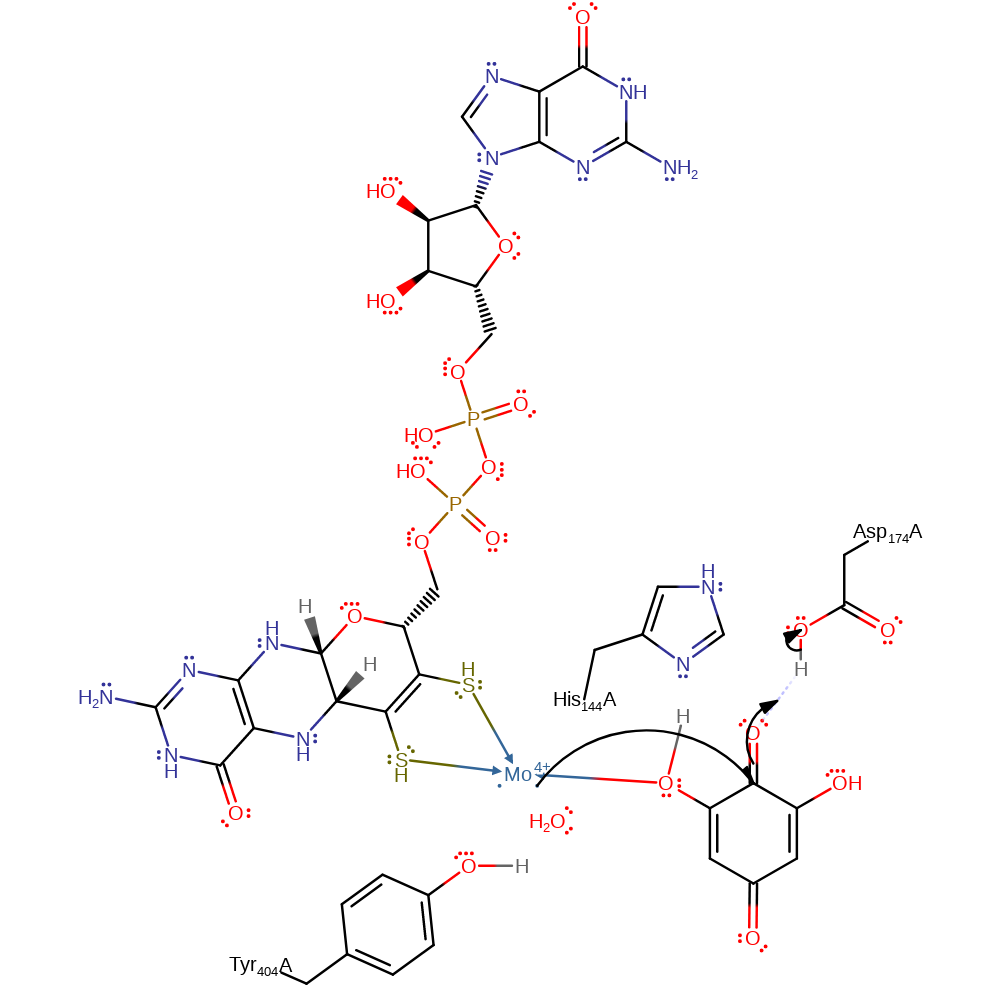
Step 8. Mo(IV) donates a single electron into the bound quinone, resulting in the protonation of the ketone group at the 2 position from Asp174.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His144A | hydrogen bond acceptor |
| Asp174A | hydrogen bond donor |
| Tyr404A | hydrogen bond donor |
| Asp174A | proton donor |
Chemical Components
redox reaction, proton transfer, intermediate formation, inferred reaction step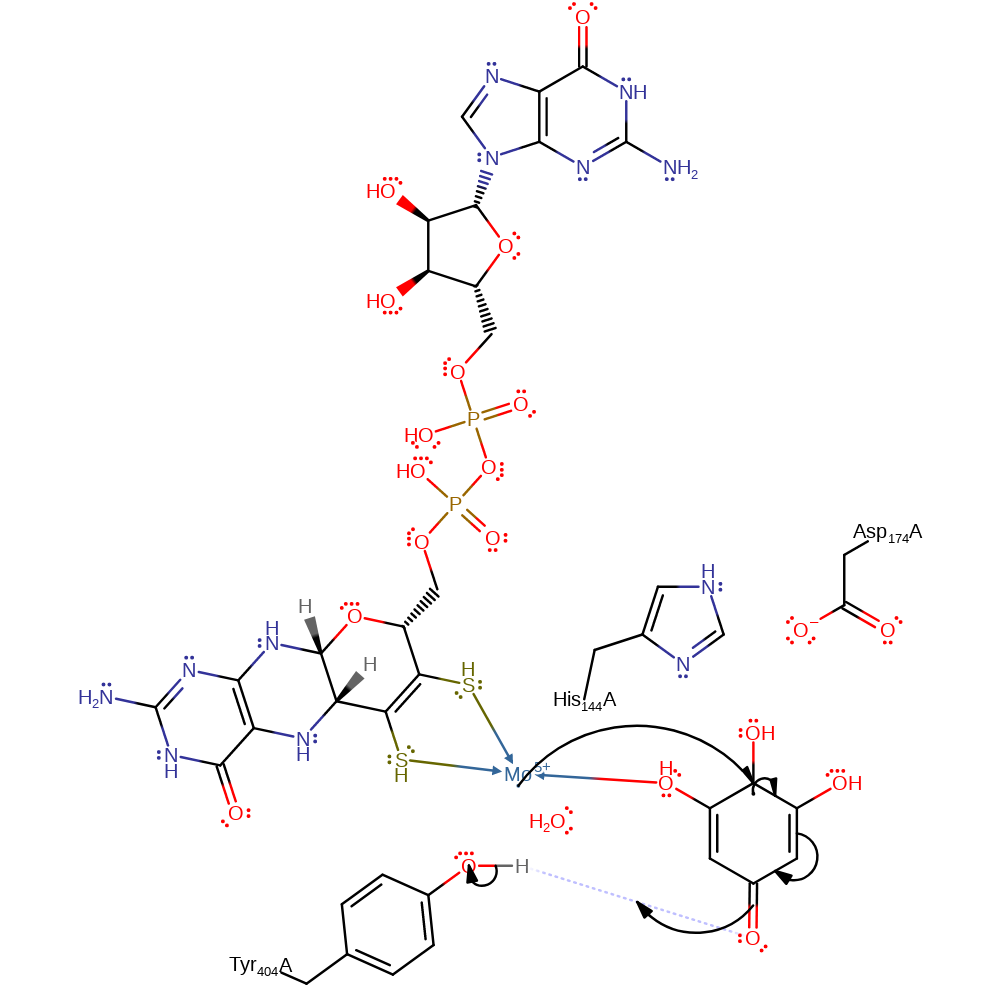
Step 9. Mo(V) donates a second electron to the bound quinone, causing the ketone at the 5 position to protonate from Tyr404. The 1,2,3,5-tetrahydroxybenzene is displaced from the molybdenum's coordination sphere by water.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His144A | hydrogen bond acceptor |
| Asp174A | hydrogen bond acceptor |
| Tyr404A | hydrogen bond donor |
| Tyr404A | proton donor |
Chemical Components
ingold: bimolecular nucleophilic substitution, electron transfer, proton transfer, radical termination, coordination to a metal ion, decoordination from a metal ion, intermediate terminated, overall product formed, native state of cofactor regenerated, native state of enzyme regenerated, inferred reaction stepIntroduction
This mechanism illustrates a transhydroxylase reaction via `Umpolung' of the substrate by oxidation to an ortho-quinone and nucleophilic hydroxylation by a molybdenum-coordinated OH group. Firstly Pyrogallol converts the C2 and C3 hydroxyls into ketone groups. Then the Molybdenum coordinated OH attacks the C5 carbon which results in the feeding of electrons to the C3 O which will accept a proton. Next C5 carbon is deprotonated and this results in another rearrangemnt of double bonds and the feeding of electrons to the C2 O which will also accept a proton. The C5 hydroxyl will then decoordinate from the Molybdenum ion and the tetrahydroxybenzene intermediate that has been formed will rotate 180 degrees in the active site and will react in the reverse way than before which is characteristic of an "Umpolung" mechanism.
Catalytic Residues Roles
| UniProt | PDB* (4v4e) |
Chemical Components
hydride transfer, proton transfer, aromatic unimolecular elimination by the conjugate base, intermediate formation, overall reactant used, bimolecular nucleophilic addition, cofactor used, decoordination from a metal ion, coordination to a metal ion, aromatic bimolecular electrophilic addition, intermediate collapse, native state of cofactor regenerated, overall product formedReferences
- Messerschmidt A et al. (2004), Proc Natl Acad Sci U S A, 101, 11571-11576. Crystal structure of pyrogallol-phloroglucinol transhydroxylase, an Mo enzyme capable of intermolecular hydroxyl transfer between phenols. DOI:10.1073/pnas.0404378101. PMID:15284442.
- Hille R et al. (1998), FEMS Microbiol Rev, 22, 489-501. Mechanistic aspects of molybdenum-containing enzymes. DOI:10.1016/s0168-6445(98)00037-0. PMID:10189201.

Step 1. Pyrogallol converts itself to 3-Hydroxy-o-benzochinon by deprotonating the C2 hydroxyl which then eliminates the hydride on the C3 hydroxyl.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
hydride transfer, proton transfer, ingold: aromatic unimolecular elimination by the conjugate base, intermediate formation, overall reactant used
Step 2. The hydroxyl group coordinated to Mo nucleophilically attacks C5 which results in the rearrangement of electrons in the aromatic ring and as a result the C3 oxygen can accept a proton.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
ingold: bimolecular nucleophilic addition, cofactor used, proton transfer, intermediate formation
Step 3. C5 is deprotonated which results in another rearrangement of double bonds and as a result the C2 oxygen accepts a proton from the surrounding solvent.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
proton transfer, intermediate formationCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
decoordination from a metal ion
Step 5. The tetrahydroxybenzene intermediate will turn 180 degrees at the active site and react with the molybdenum centre in a reverse way as described in the first half of the cycle
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
Catalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
coordination to a metal ion, cofactor used, intermediate formation
Step 7. The C5 hyrdroxyl releases a proton to the surrounding solution which results 9n the ffeeding of electrons around the aromati ring where then the double bond between C1 and C2 accepts a proton from the solvent.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
ingold: aromatic bimolecular electrophilic addition, proton transfer, intermediate formation
Step 8. The C2 hydroxyl is cleaved from the aromatic ring and the electrons are fed back round the ring to the C5 oxygen which then accepts a proton from the surrounding solvent.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
ingold: aromatic unimolecular elimination by the conjugate base, decoordination from a metal ion, intermediate collapse, native state of cofactor regenerated, overall product formedIntroduction
This proposal follows a mechanism which goes via an "Umpolung" of the hydroxyl group by the molybdenum cofactor. This includes an electrophilic substitution by the Mo-oxo group which occurs at position 5 of pyrogallol. In a series of proton transfers to and from the surrounding solvent to oxygen groups on C2 and C5; a hydroxyl group originating from the Mo-oxo group is added onto the C5 carbon and the hydroxyl group on C2 is removed to produce the final product of phloroglucinol.
Catalytic Residues Roles
| UniProt | PDB* (4v4e) |
Chemical Components
bimolecular electrophilic addition, coordination to a metal ion, enzyme-substrate complex formation, intermediate formation, overall reactant used, proton transfer, electron transfer, decoordination from a metal ion, enzyme-substrate complex cleavage, hydride transfer, inferred reaction step, aromatic unimolecular elimination by the conjugate base, intermediate collapse, overall product formedReferences
- Messerschmidt A et al. (2004), Proc Natl Acad Sci U S A, 101, 11571-11576. Crystal structure of pyrogallol-phloroglucinol transhydroxylase, an Mo enzyme capable of intermolecular hydroxyl transfer between phenols. DOI:10.1073/pnas.0404378101. PMID:15284442.
- Hille R et al. (1998), FEMS Microbiol Rev, 22, 489-501. Mechanistic aspects of molybdenum-containing enzymes. DOI:10.1016/s0168-6445(98)00037-0. PMID:10189201.

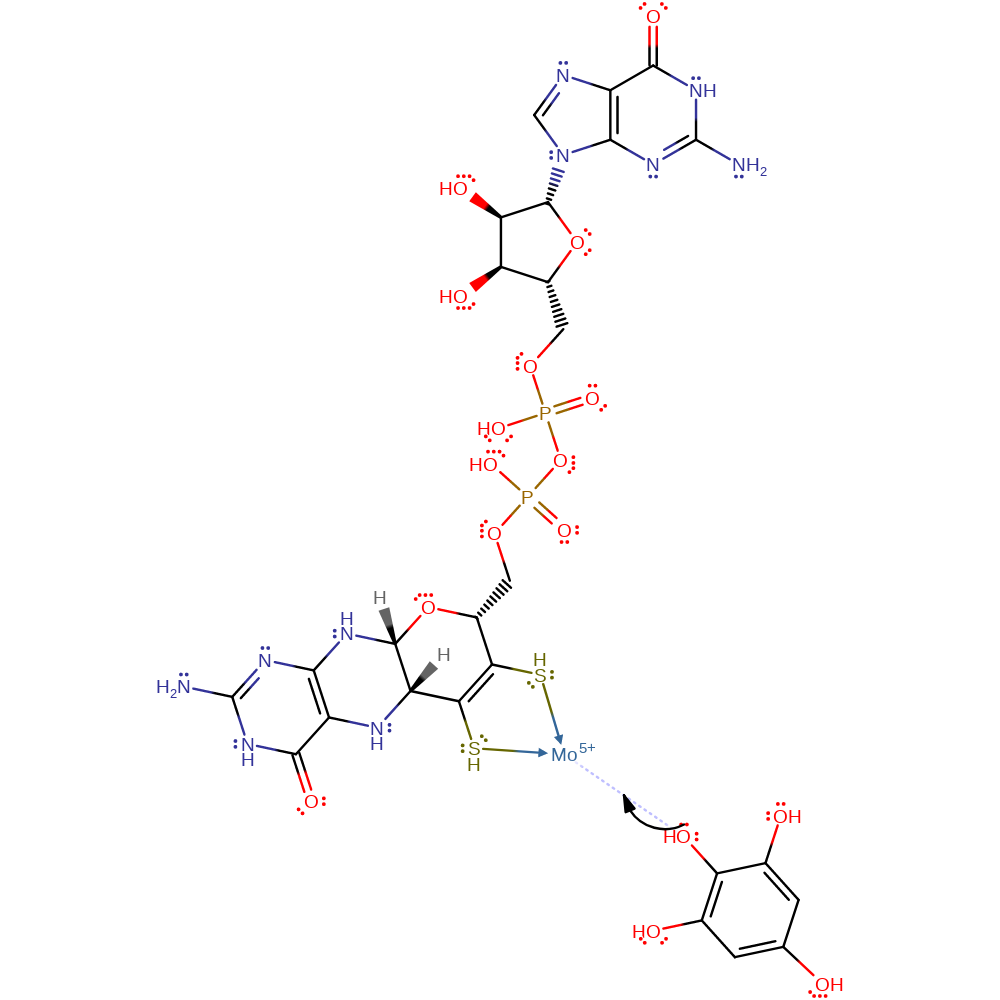
Step 1. A proton from the C2 hydroxyl is lost to the solution which results in the movement of electrons through double bond arrangements around the aromatic ring. The by electrophilic addition the C4-C5 bond accepts the Mo-oxo group.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
ingold: bimolecular electrophilic addition, coordination to a metal ion, enzyme-substrate complex formation, intermediate formation, overall reactant used
Step 2. The C5 hydrogen is released which results in the rearrangement of double bonds which feed electrons to the C2 oxygen which then accepts a proton from the surrounding solvent.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
proton transfer, electron transfer, intermediate formation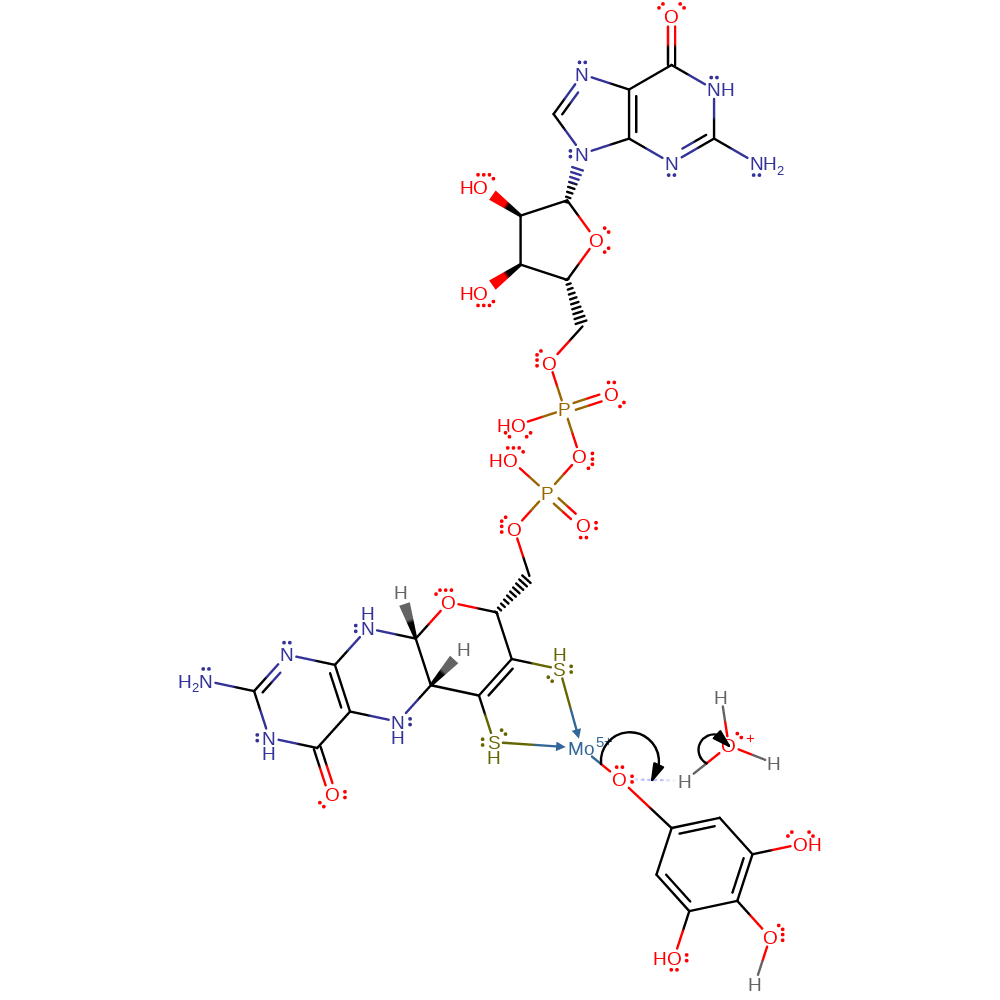
Step 3. The Mo coordinated oxygen accepts a proton which results in the decoordination from the metal ion.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
proton transfer, decoordination from a metal ion, enzyme-substrate complex cleavage, intermediate formation
Step 4. The pyrogallol intermediate loses 2 electrons resulting in forming 2 keto groups on the C2 and C5 carbon. Meanwhile the molybdenum ion accepts 2 electrons thus going from Mo(VI) to Mo (IV). Also the intermediate substrate rotates in the active site so that the C2 hydroxyl can now interact with the cofactor.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
electron transfer, hydride transfer, intermediate formation, inferred reaction step, proton transfer
Step 5. Mo(IV) coordinates to the oxygen at C2 via the lone pair of electrons it gained in the previous step. This results in a series of electron transfers around the aromatic ring to the C5 oxygen which will then accept a proton
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
coordination to a metal ion, electron transfer, intermediate formation, proton transfer, enzyme-substrate complex formation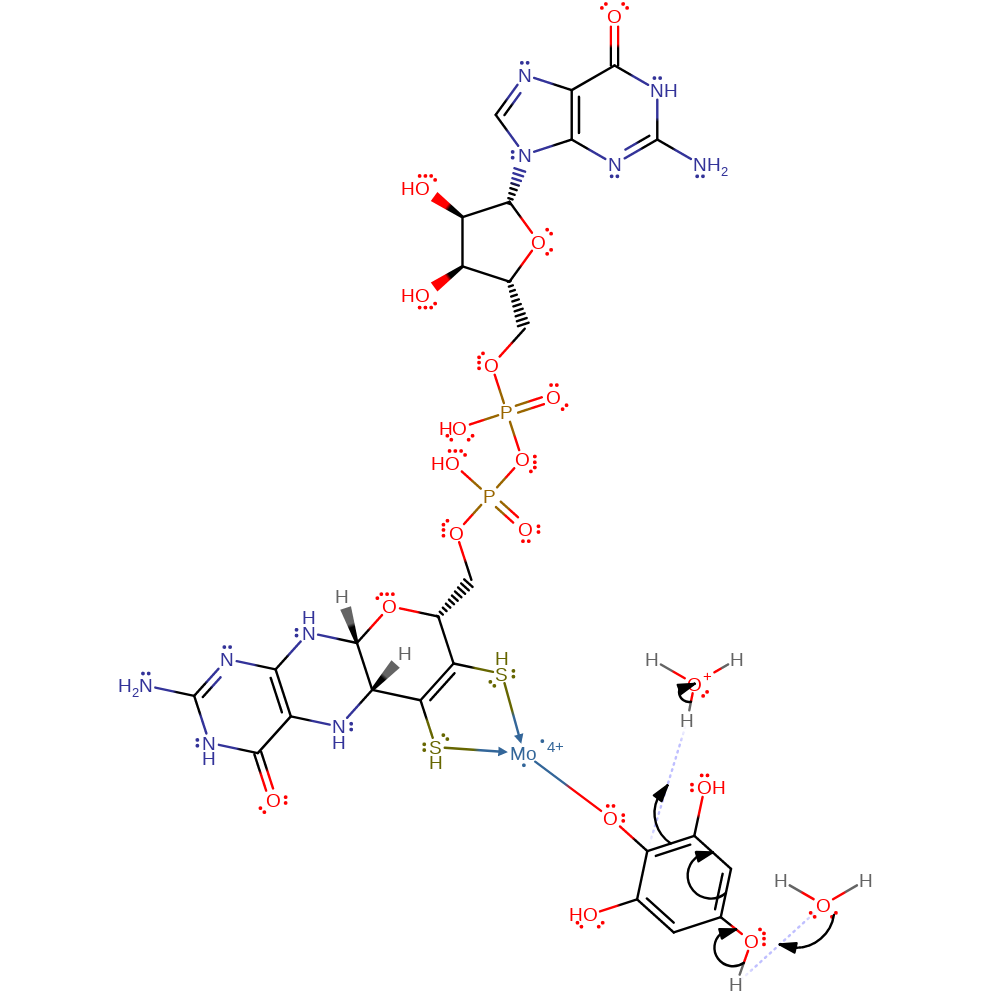
Step 6. The C5 hydroxyl releases a proton and transfer electrons back round the aromatic ring where the C2-C3 double bond will accept a proton from the solvent.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
ingold: bimolecular electrophilic addition, intermediate formation, proton transfer, electron transfer
Step 7. The coordinate bond between the substrate and Molybdenum breaks which results in the cleavage of the C2 hydroxyl group and the transfer of electrons around the aromatic ring to the C5 oxygen which will then accept a proton producing phloroglucinol.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|



 Download:
Download: 
 Download:
Download:  Download:
Download: