Transaldolase (type I)
Transaldolase (TAL) is an enzyme of the pentose phosphate pathway (PPP) found almost ubiquitously in the three domains of life (Archaea, Bacteria, and Eukarya). It is important for the balance of metabolites in the pentose-phosphate pathway, and catalyses the reversible transfer of a three-carbon ketol unit from sedoheptulose 7-phosphate to glyceraldehyde 3-phosphate to form erythrose 4-phosphate and fructose 6-phosphate. This enzyme, together with transketolase, provides a link between the glycolytic and pentose-phosphate pathways.
Reference Protein and Structure
- Sequence
-
P0A870
 (2.2.1.2)
(2.2.1.2)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1onr
- STRUCTURE OF TRANSALDOLASE B
(1.87 Å)



- Catalytic CATH Domains
-
3.20.20.70
 (see all for 1onr)
(see all for 1onr)
- Cofactors
- Water (1)
Enzyme Reaction (EC:2.2.1.2)
Enzyme Mechanism
Introduction
In this proposal the Asp 17 is no longer taking part in the catalytic mechanism, instead Glu 96 takes its role as general acid/base. Firstly Glu96 actS as a general base and deprotonates the conserved Lys132 in TalB via the catalytic water molecule to facilitate the nucleophilic attack of the NH2 group on the carbonyl carbon atom of Fructose 6-phosphate. The nucleophilic attack is followed by a protonation of the di-polar alkoxide in which the NH2R+ group serves as a proton source. Next, the Glu–H2O dyad acts as a general acid in the protonation of the C2-OH group of the neutral carbinolamine intermediate. Finally, a water molecule is released and the Schiff base intermediate is formed. Then Glu 96 deprotonates a water so that it can act as a base which will deprotonate the C4-OH and results in the heterolytic cleavage and the release of D-glyceraldehyde 3-phosphate. The DHA moiety is still bound to the lysyl residue as a carbanion which will undergo an isomerisation in which an electron pair will be transferred to the Nitrogen of Lys 132. The Lysine bound intermediate attacks the carbonyl carbon of D-erythrose 4-phosphate and the oxyanion formed will deprotonate Glu 96. Glu 96 then deprotonates a water which will attack the carabamylated carbon of the intermediate. Lastly Lys 132 deprotonates a bound waterwhich will initiate an elimination and result in the release of D-sedoheptulose 7-phosphate product from the enzyme. The active site is regenerated via protonation of Lys 132 by a bound water which in turn will accept a proton from Glu 96.
Catalytic Residues Roles
| UniProt | PDB* (1onr) | ||
| Phe178 | Phe178(177)A | Prevents Fructose 6-P aldolase actvity as cannot form a hydrogen bond to the water which allows it to participate in the reaction. | steric role |
| Glu96 | Glu96(95)A | Acts as a general acid/base and relays protons to and from the catalytic water enabling it to attack the substrate. | proton acceptor, electrostatic stabiliser, proton donor |
| Lys132 | Lys132(131)A | Acts as a general acid/base, and also as the catalytic nucleophile. | covalently attached, nucleofuge, nucleophile, proton acceptor, proton donor, electron pair acceptor, electron pair donor |
| Thr156 | Thr156(155)A | Stabilises catalytic water in active site by forming hydrogen bonds with it | electrostatic stabiliser |
Chemical Components
proton transfer, overall reactant used, bimolecular nucleophilic addition, inferred reaction step, intermediate formation, enzyme-substrate complex formation, unimolecular elimination by the conjugate base, schiff base formed, intermediate collapse, overall product formed, tautomerisation (not keto-enol), michael addition, proton relay, bimolecular elimination, enzyme-substrate complex cleavage, intermediate terminated, native state of enzyme regeneratedReferences
- Stellmacher L et al. (2015), ChemCatChem, 7, 3140-3151. Acid-Base Catalyst Discriminates between a Fructose 6-Phosphate Aldolase and a Transaldolase. DOI:10.1002/cctc.201500478.
- Lehwess-Litzmann A et al. (2011), Nat Chem Biol, 7, 678-684. Twisted Schiff base intermediates and substrate locale revise transaldolase mechanism. DOI:10.1038/nchembio.633. PMID:21857661.

Step 1. Glu96 deprotonatesa water which in turn accepts a proton from Lys132.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr156(155)A | electrostatic stabiliser |
| Phe178(177)A | steric role |
| Lys132(131)A | proton donor |
| Glu96(95)A | proton acceptor |
Chemical Components
proton transfer, overall reactant used
Step 2. Lys132 nucleophilically attacks the carbonyl carbon meanwhile the oxyanion produced is also protonated by Lys132
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu96(95)A | electrostatic stabiliser |
| Phe178(177)A | steric role |
| Lys132(131)A | covalently attached, proton donor, nucleophile |
Chemical Components
ingold: bimolecular nucleophilic addition, inferred reaction step, intermediate formation, enzyme-substrate complex formation, proton transfer, overall reactant used
Step 3. To form the Fructose 6-phosphate Schiff base which elimates the hydroxyl group which will then accept a proton from the catalytic water which will then accepts a proton from Glu96.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Phe178(177)A | steric role |
| Thr156(155)A | electrostatic stabiliser |
| Lys132(131)A | covalently attached |
| Glu96(95)A | proton donor |
| Lys132(131)A | electron pair donor |
Chemical Components
ingold: unimolecular elimination by the conjugate base, proton transfer, intermediate formation, schiff base formed
Step 4. Glu96 deprotonates water so that it can act as a base which deprotonates the C4-OH and results in the heterolytic cleavage and the release of the first product, G3P.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr156(155)A | electrostatic stabiliser |
| Phe178(177)A | steric role |
| Lys132(131)A | covalently attached |
| Glu96(95)A | proton acceptor |
Chemical Components
proton transfer, ingold: unimolecular elimination by the conjugate base, intermediate collapse, overall product formed
Step 5. The carbanion undergoes an isomerisation in which the lone pair of electrons migrate to the nitrogen of Lys132.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys132(131)A | covalently attached |
| Thr156(155)A | electrostatic stabiliser |
| Phe178(177)A | steric role |
| Lys132(131)A | electron pair acceptor |
Chemical Components
tautomerisation (not keto-enol), intermediate formation
Step 6. The lysine-bound intermediate attacks the carbonyl carbon of the D-erythrose 4-phosphate substrate in a 1,4-nucleophilic addition. The formed oxyanion deprotonates Glu96.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys132(131)A | covalently attached |
| Thr156(155)A | electrostatic stabiliser |
| Phe178(177)A | steric role |
| Glu96(95)A | proton donor |
| Lys132(131)A | electron pair donor |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer, overall reactant used, enzyme-substrate complex formation, intermediate formation, michael addition
Step 7. Glu96 deprotonates a water molecule, which attacks the carbamylated carbon of the intermediate in a nucleophilic addition.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr156(155)A | electrostatic stabiliser |
| Phe178(177)A | steric role |
| Glu96(95)A | proton acceptor |
| Lys132(131)A | electron pair acceptor |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer, intermediate formation, proton relay
Step 8. Lys132 deprotonates the bound water, initiating an elimination reaction that releases the D-sedoheptulose 7-phosphate product from the enzyme.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr156(155)A | electrostatic stabiliser |
| Phe178(177)A | steric role |
| Lys132(131)A | proton acceptor, nucleofuge |
Chemical Components
ingold: bimolecular elimination, enzyme-substrate complex cleavage, overall product formed, proton relay, intermediate collapse, intermediate terminated
Step 9. Regeneration of the active site in which the Lys132 deprotonates a bound water that in turn deprotonates Glu96.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys132(131)A | proton acceptor |
| Glu96(95)A | proton donor |
Chemical Components
proton transfer, proton relay, native state of enzyme regeneratedIntroduction
Glu96 activates the catalytic lysine, which attacks the carbonyl carbon of D-fructose 6-phosphate in a nucleophilic addition. The generated oxyanion is reprotonated from water, which in turn deprotonates the now bound lysine. Lys132 initiates an elimination of the hydroxide group, which deprotonates the Glu96 to form water. Asp17 deprotonates the hydroxide group gamma to the carbamylated carbon, initiating a bimolecular elimination of the product D-glyceraldehyde 3-phosphate from the covalently bound intermediate and resulting in a carbanion. The carbanion undergoes an isomerisation in which the lone pair of electrons migrate to the nitrogen of Lys132. The final steps of this mechanism are the reverse of the first half of the reaction [PMID:11298760]. There is a crystallographic water molecule between Glu96 and Lys132 which is involved in catalysis.
Catalytic Residues Roles
| UniProt | PDB* (1onr) | ||
| Asp17, Glu96 | Asp17(16)A, Glu96(95)A | Acts as a general acid/base. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Lys132 | Lys132(131)A | Acts as a general acid/base, and also as the catalytic nucleophile. | covalently attached, hydrogen bond acceptor, hydrogen bond donor, nucleofuge, proton acceptor, proton donor, nucleophile, electron pair acceptor, electron pair donor |
| Thr156 | Thr156(155)A | Located close to the C2 carbon atom of the donor substrate and this conserved side chain might provide additional catalytic power through stabilisation of the negative charge developing at the C2 oxygen atom in the transition state of the formation of the carbinolamine | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
proton transfer, proton relay, bimolecular nucleophilic addition, overall reactant used, enzyme-substrate complex formation, intermediate formation, unimolecular elimination by the conjugate base, enzyme-substrate complex cleavage, intermediate collapse, dehydration, schiff base formed, bimolecular elimination, overall product formed, tautomerisation (not keto-enol), michael addition, intermediate terminated, native state of enzyme regeneratedReferences
- Schörken U et al. (2001), Eur J Biochem, 268, 2408-2415. Identification of catalytically important residues in the active site ofEscherichia colitransaldolase. DOI:10.1046/j.1432-1327.2001.02128.x. PMID:11298760.

Step 1. Initial activation of Lys132 in which Glu96 deprotonates a bound water that in turn deprotonates Lys132.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu96(95)A | hydrogen bond acceptor |
| Thr156(155)A | hydrogen bond donor |
| Lys132(131)A | hydrogen bond donor |
| Asp17(16)A | hydrogen bond acceptor |
| Lys132(131)A | proton donor |
| Glu96(95)A | proton acceptor |
Chemical Components
proton transfer, proton relay
Step 2. Lys132 attacks the carbonyl carbon of D-fructose 6-phosphate in a nucleophilic addition. The generated oxyanion is reprotonated from water, which in turn deprotonates the now bound lysine
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu96(95)A | hydrogen bond donor |
| Thr156(155)A | hydrogen bond donor, electrostatic stabiliser |
| Lys132(131)A | hydrogen bond donor |
| Asp17(16)A | hydrogen bond acceptor |
| Lys132(131)A | proton donor, nucleophile |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, overall reactant used, enzyme-substrate complex formation, proton relay, intermediate formation
Step 3. Lys132 initiates an elimination of the hydroxide group, which deprotonates the Glu96 to form water.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu96(95)A | hydrogen bond donor |
| Thr156(155)A | hydrogen bond donor |
| Lys132(131)A | covalently attached |
| Asp17(16)A | hydrogen bond acceptor |
| Glu96(95)A | proton donor |
| Lys132(131)A | electron pair donor |
Chemical Components
ingold: unimolecular elimination by the conjugate base, proton transfer, enzyme-substrate complex cleavage, proton relay, intermediate formation, intermediate collapse, dehydration, schiff base formed
Step 4. Asp17 deprotonates the hydroxide group gamma to the carbamylated carbon, initiating a bimolecular elimination of the product D-glyceraldehyde 3-phosphate from the covalently bound intermediate and resulting in a carbanion.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu96(95)A | hydrogen bond acceptor |
| Lys132(131)A | covalently attached |
| Asp17(16)A | hydrogen bond acceptor, proton acceptor |
Chemical Components
ingold: bimolecular elimination, enzyme-substrate complex cleavage, intermediate collapse, intermediate formation, overall product formed
Step 5. The carbanion undergoes an isomerisation in which the lone pair of electrons migrate to the nitrogen of Lys132.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu96(95)A | hydrogen bond acceptor |
| Lys132(131)A | covalently attached |
| Asp17(16)A | hydrogen bond donor |
| Lys132(131)A | electron pair acceptor |
Chemical Components
tautomerisation (not keto-enol), intermediate formation
Step 6. The lysine-bound intermediate attacks the carbonyl carbon of the D-erythrose 4-phosphate substrate in a 1,4-nucleophilic addition. The formed oxyanion deprotonates Asp17.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu96(95)A | hydrogen bond acceptor |
| Thr156(155)A | hydrogen bond donor |
| Lys132(131)A | covalently attached |
| Asp17(16)A | hydrogen bond donor, proton donor |
| Lys132(131)A | electron pair donor |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer, overall reactant used, enzyme-substrate complex formation, intermediate formation, michael addition
Step 7. Glu96 deprotonates a water molecule, which attacks the carbamylated carbon of the intermediate in a nucleophilic addition.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu96(95)A | hydrogen bond acceptor |
| Thr156(155)A | hydrogen bond donor, electrostatic stabiliser |
| Lys132(131)A | covalently attached |
| Asp17(16)A | hydrogen bond acceptor |
| Glu96(95)A | proton acceptor |
| Lys132(131)A | electron pair acceptor |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer, enzyme-substrate complex formation, intermediate formation, proton relay
Step 8. Lys132 deprotonates the bound water, initiating an elimination reaction that releases the D-sedoheptulose 7-phosphate product from the enzyme.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu96(95)A | hydrogen bond donor |
| Thr156(155)A | hydrogen bond donor |
| Lys132(131)A | hydrogen bond acceptor |
| Asp17(16)A | hydrogen bond acceptor |
| Lys132(131)A | nucleofuge, proton acceptor |
Chemical Components
ingold: bimolecular elimination, enzyme-substrate complex cleavage, overall product formed, proton relay, intermediate collapse, intermediate terminated
Step 9. Regeneration of the active site in which the Lys132 deprotonates a bound water that in turn deprotonates Glu96.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu96(95)A | hydrogen bond donor |
| Thr156(155)A | hydrogen bond donor |
| Lys132(131)A | hydrogen bond acceptor |
| Asp17(16)A | hydrogen bond acceptor |
| Glu96(95)A | proton donor |
| Lys132(131)A | proton acceptor |


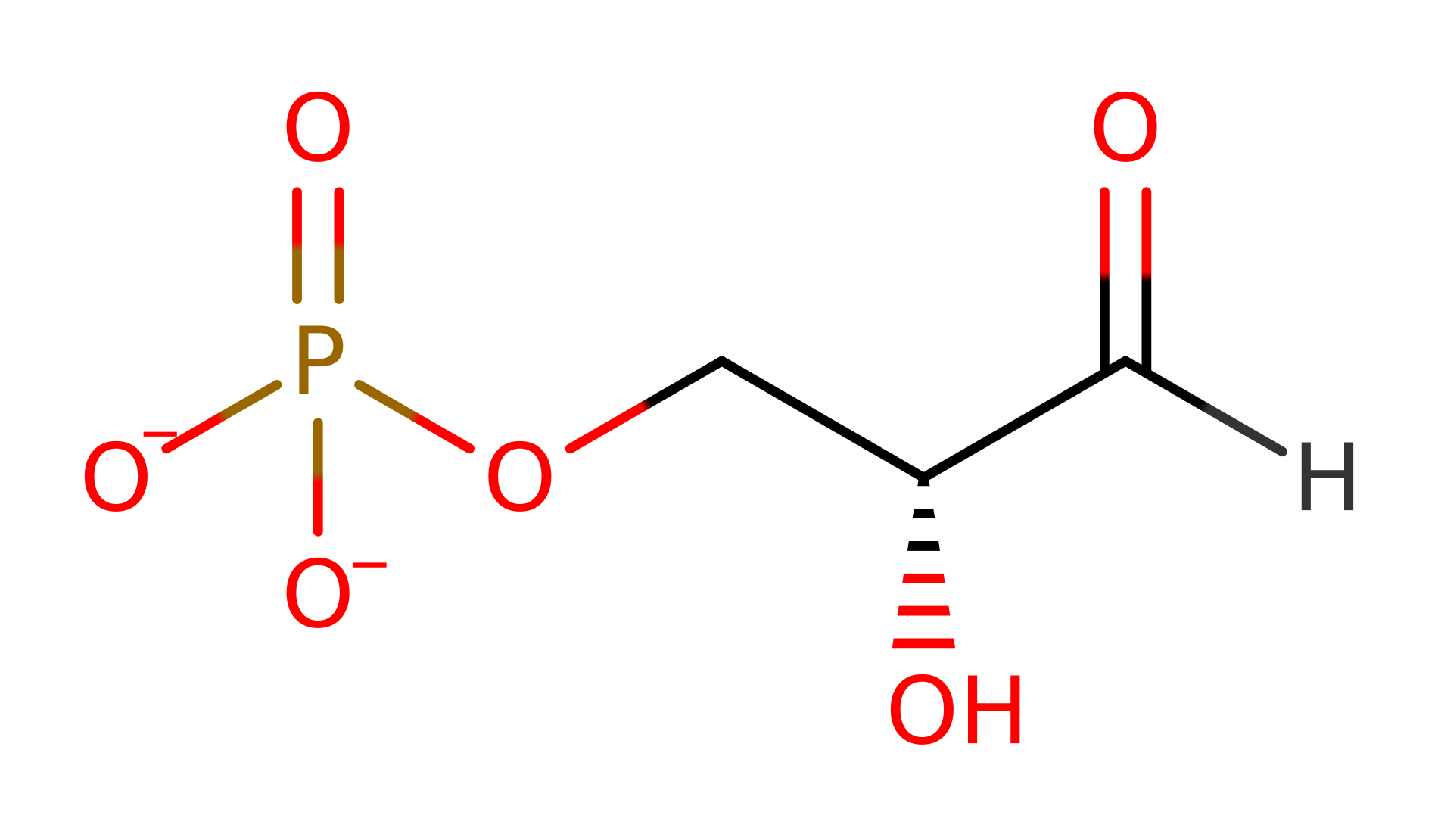

 Download:
Download:  Download:
Download: