Cerebroside-sulfatase
Sulphatases are an evolutionarily highly conserved gene family. They hydrolyse sulphate ester bonds in a wide variety of structurally different compounds ranging from complex glucosaminoglucans and glycolipids to sulphated hydroxysteroids and amino acids. Among the sulphatase family, lysosomal arylsulphatase A (ASA) has been most extensively studied.
The major physiological substrate of human ASA is a sphingolipid sulphate ester, cerebroside 3-sulphate, a major constituent of the myelin sheet. In vivo, the 3-sulphate group can be hydrolysed by ASA only if cerebroside 3-sulphate is complexed with the small activator protein saposin B solubilising the hydrophobic substrate.
Reference Protein and Structure
- Sequence
-
P15289
 (3.1.6.8)
(3.1.6.8)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Homo sapiens (Human)

- PDB
-
1auk
- HUMAN ARYLSULFATASE A
(2.1 Å)



- Catalytic CATH Domains
-
3.40.720.10
 (see all for 1auk)
(see all for 1auk)
- Cofactors
- Magnesium(2+) (1) Metal MACiE
Enzyme Reaction (EC:3.1.6.8)
Enzyme Mechanism
Introduction
ASA requires the post-translational oxidation of a cysteine sidechain to an aldehyde, yielding a formylglycine (3 letter code FGL). This is reduced to a gem-diol by a hydroxide ion. One of the alcohol groups, activated by magnesium, is deprotonated by glutamate and then carries out a nucleophilic attack on the substrate. The sulphate ester bond is broken, and the negatively charged leaving group abstracts a proton from a histidine residue. Deprotonation of the non-nucleophilic alcohol group of formylglycine leads to the aldehyde being re-formed and the enzyme-substrate bond being broken, restoring the enzyme to its original state.
Catalytic Residues Roles
| UniProt | PDB* (1auk) | ||
| Ser150 | Ser150(132)A | Withdraws electron density from the sulfate oxygen atoms, leading to an increased electrophilicity of the sulfur centre. | increase electrophilicity, hydrogen bond donor, electrostatic stabiliser |
| Arg73 | Arg73(55)A | Helps stabilise the formylglycine residue and the intermediates formed. | hydrogen bond acceptor, hydrogen bond donor, electrostatic stabiliser |
| Lys302, Lys123 | Lys302(284)A, Lys123(105)A | Withdraws electron density from the sulphate oxygen atoms, leading to an increased electrophilicity of the sulphur centre. | hydrogen bond donor, electrostatic stabiliser |
| His125 | His125(107)A | Activates formylglycine to encourage the gem-diol form and protonates it. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, electrostatic stabiliser |
| Cys69 (ptm) | Fgl69(51)A (ptm) | Forms part of the metal binding site. Acts as a catalytic nucleophile, activated by Asp281, it attacks the sulfur atom of the sulfate group, eliminating the cerebroside product and forming a sulfated formylglycine residue. formylglycine is regenerated via base activate elimination of the sulfate group, followed by addition of water to the carbonyl group. | hydrogen bond acceptor, hydrogen bond donor, nucleophile, metal ligand, proton acceptor, proton donor, proton relay, electrostatic stabiliser, electrofuge, electrophile |
| His229 | His229(211)A | Protonates the alcohol leaving group, deprotonates the water molecule which converts formylglycine to the gem-diol form. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, electrostatic stabiliser |
| Asp281 | Asp281(263)A | Forms part of the metal binding site. Deprotonates the gem-diol form of the nucleophilic residue to activate it. | hydrogen bond acceptor, hydrogen bond donor, metal ligand, proton acceptor, proton donor |
| Asn282, Asp29, Asp30 | Asn282(264)A, Asp29(11)A, Asp30(12)A | Forms part of the metal binding site. | metal ligand |
Chemical Components
bimolecular nucleophilic substitution, proton transfer, overall reactant used, enzyme-substrate complex formation, intermediate formation, overall product formed, bimolecular elimination, enzyme-substrate complex cleavage, intermediate collapse, bimolecular nucleophilic addition, intermediate terminated, native state of enzyme regeneratedReferences
- von Bülow R et al. (2001), J Mol Biol, 305, 269-277. Crystal Structure of an Enzyme-Substrate Complex Provides Insight into the Interaction between Human Arylsulfatase A and its Substrates During Catalysis. DOI:10.1006/jmbi.2000.4297. PMID:11124905.
- Ghosh D (2007), Cell Mol Life Sci, 64, 2013-2022. Human sulfatases: A structural perspective to catalysis. DOI:10.1007/s00018-007-7175-y. PMID:17558559.
- Chruszcz, Maksymilian et al. (2003), J. Inorg. Biochem., 96, 386-392. Crystal structure of a covalent intermediate of endogenous human arylsulfatase A. PMID:12888274.
- Lukatela G et al. (1998), Biochemistry, 37, 3654-3664. Crystal Structure of Human Arylsulfatase A: The Aldehyde Function and the Metal Ion at the Active Site Suggest a Novel Mechanism for Sulfate Ester Hydrolysis†,‡. DOI:10.1021/bi9714924. PMID:9521684.
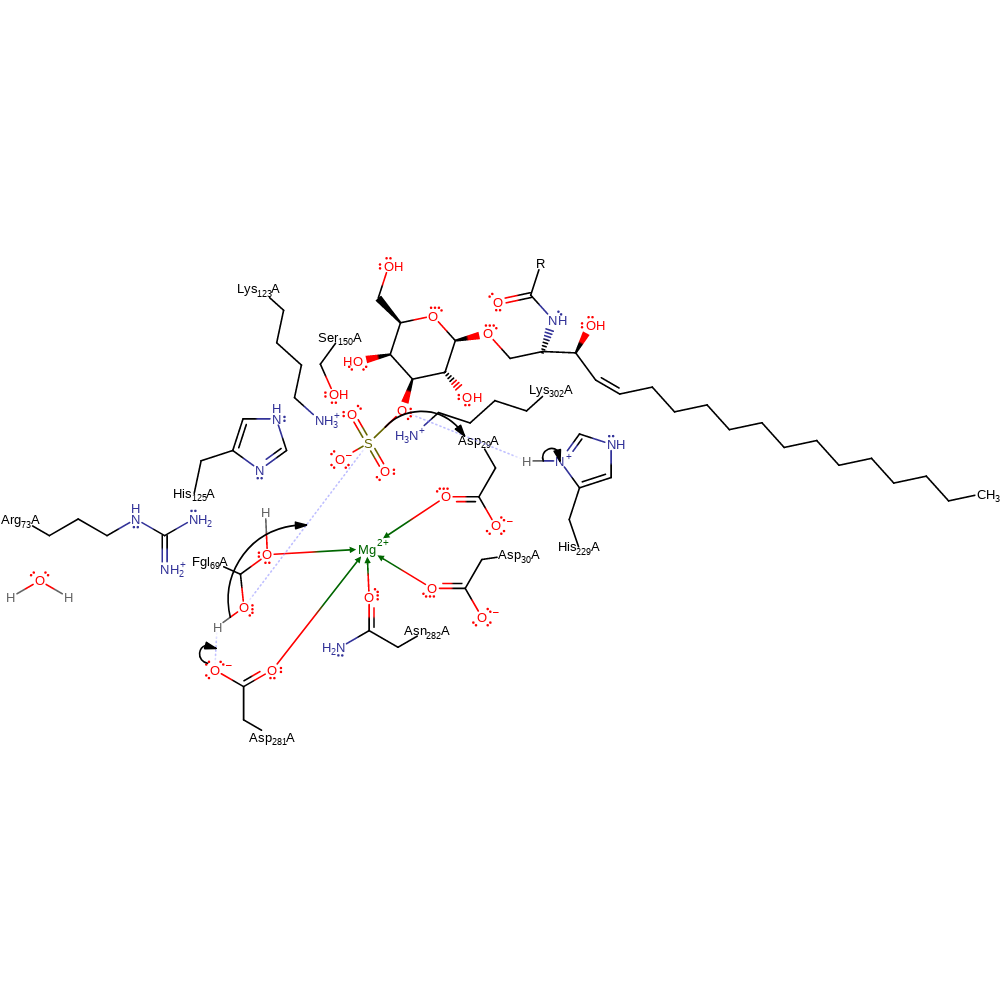
Step 1. Asp281 deprotonates the post-translationally modified formylglycine, which attacks the sulfate of the cerebroside 3-sulfate substrate in a nucleophilic substitution, releasing the cerebroside product which is reprotonated from His229.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys123(105)A | electrostatic stabiliser, hydrogen bond donor, increase electrophilicity |
| Ser150(132)A | electrostatic stabiliser, hydrogen bond donor, increase electrophilicity |
| His125(107)A | hydrogen bond acceptor, electrostatic stabiliser |
| Lys302(284)A | hydrogen bond donor, electrostatic stabiliser |
| Fgl69(51)A (ptm) | hydrogen bond donor, hydrogen bond acceptor, nucleophile |
| Asp281(263)A | hydrogen bond acceptor |
| His229(211)A | hydrogen bond donor, electrostatic stabiliser |
| Arg73(55)A | hydrogen bond donor, electrostatic stabiliser |
| Asp29(11)A | metal ligand |
| Asp30(12)A | metal ligand |
| Asp281(263)A | metal ligand |
| Asn282(264)A | metal ligand |
| Fgl69(51)A (ptm) | metal ligand |
| Fgl69(51)A (ptm) | proton donor |
| Asp281(263)A | proton acceptor |
| His229(211)A | proton donor |
Chemical Components
ingold: bimolecular nucleophilic substitution, proton transfer, overall reactant used, enzyme-substrate complex formation, intermediate formation, overall product formed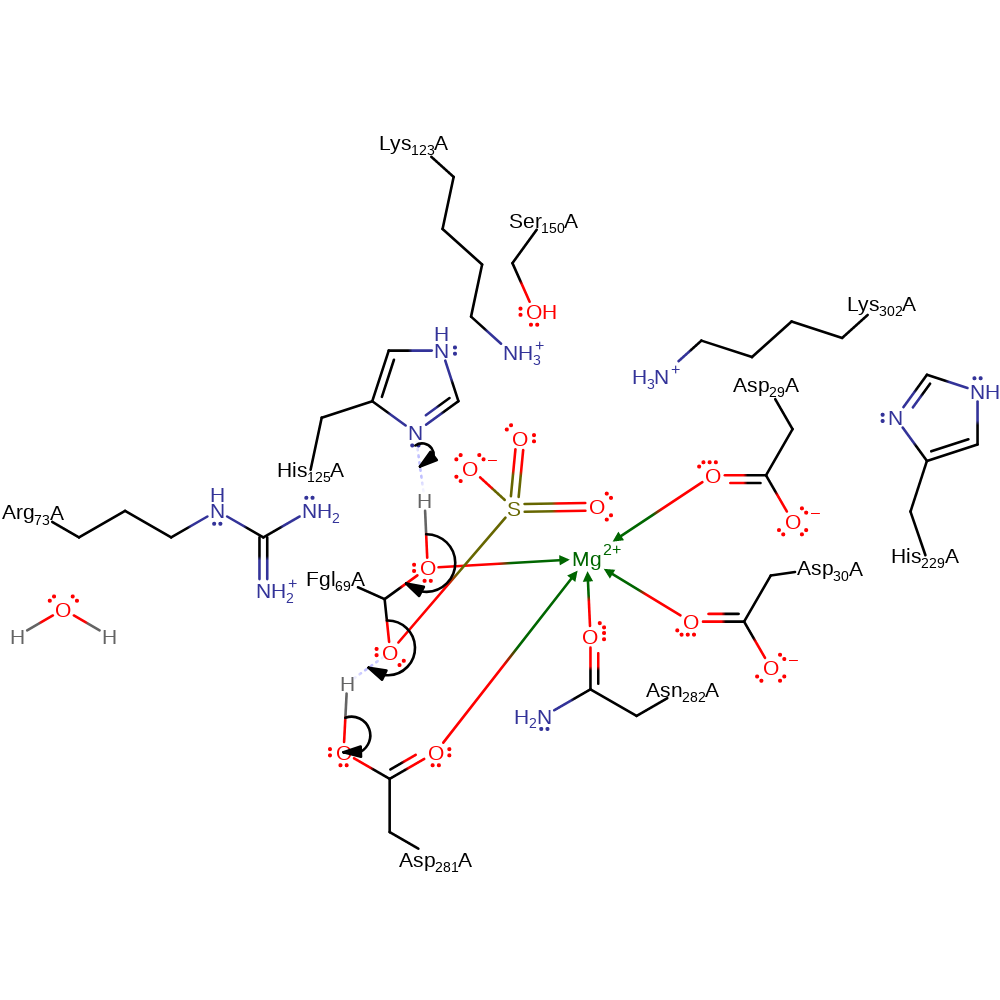
Step 2. His125 deprotonates the other alcohol group of the formylglycine, initiating an elimination reaction that produces the sulfate product, which is reprotonated from Asp281.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys123(105)A | electrostatic stabiliser, hydrogen bond donor |
| Ser150(132)A | electrostatic stabiliser, hydrogen bond donor |
| His125(107)A | hydrogen bond acceptor |
| Lys302(284)A | hydrogen bond donor, electrostatic stabiliser |
| Fgl69(51)A (ptm) | electrofuge, hydrogen bond acceptor, hydrogen bond donor |
| Asp281(263)A | hydrogen bond donor |
| Arg73(55)A | hydrogen bond donor, electrostatic stabiliser |
| Asp29(11)A | metal ligand |
| Asp30(12)A | metal ligand |
| Asp281(263)A | metal ligand |
| Asn282(264)A | metal ligand |
| Fgl69(51)A (ptm) | metal ligand |
| Fgl69(51)A (ptm) | proton acceptor, proton donor, proton relay |
| His125(107)A | proton acceptor |
| Asp281(263)A | proton donor |
Chemical Components
proton transfer, ingold: bimolecular elimination, enzyme-substrate complex cleavage, intermediate collapse, intermediate formation, overall product formed
Step 3. His229 deprotonates a water molecule that attacks the carbonyl carbon of the formylglycine in a nucleophilic addition, the oxyanion is reprotonated by His125.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His125(107)A | hydrogen bond donor |
| Fgl69(51)A (ptm) | hydrogen bond acceptor, electrophile, electrostatic stabiliser |
| His229(211)A | hydrogen bond acceptor |
| Arg73(55)A | hydrogen bond acceptor |
| Asp29(11)A | metal ligand |
| Asp30(12)A | metal ligand |
| Asp281(263)A | metal ligand |
| Asn282(264)A | metal ligand |
| Fgl69(51)A (ptm) | metal ligand, proton acceptor |
| His125(107)A | proton donor |
| His229(211)A | proton acceptor |





 Download:
Download: