Micrococcal nuclease
Staphyloccal nuclease is a Ca2+ activated phosphodiesterase which catalyses the hydrolysis of both DND and RNA at the 5' position of the phophodiester bond to yield a 3'-mononucleotides and polynucleotides.
Reference Protein and Structure
- Sequence
-
P00644
 (3.1.31.1)
(3.1.31.1)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Staphylococcus aureus (Bacteria)

- PDB
-
2sns
- STAPHYLOCOCCAL NUCLEASE. PROPOSED MECHANISM OF ACTION BASED ON STRUCTURE OF ENZYME-THYMIDINE 3(PRIME),5(PRIME)-BIPHOSPHATE-CALCIUM ION COMPLEX AT 1.5-ANGSTROMS RESOLUTION
(1.5 Å)



- Catalytic CATH Domains
-
2.40.50.90
 (see all for 2sns)
(see all for 2sns)
- Cofactors
- Calcium(2+) (1) Metal MACiE
Enzyme Reaction (EC:3.1.31.1)
Enzyme Mechanism
Introduction
The hydrolysis proceeds via direct nucleophilic attack on phosphate with formation of a five-coordinate, trigonal bipyramidal transition state or meta-stable intermediate followed by breakdown to from the product. The attacking nucleophile is a water molecule coordinated to the Ca2+ ion. Ca2+ facilitates the generation of hydroxide ion to allow its nucleophilic attack on the phosphate group. It also stabilises the transition state. Arg35 and Arg87 stabilise the transition state via bidentate hydrogen bonding. Arg87 is also the general acid that protonates the 5'-hydroxyl leaving group.
Catalytic Residues Roles
| UniProt | PDB* (2sns) | ||
| Asp122, Asp103, Thr123 (main-N) | Asp40A, Asp21A, Thr41A (main-N) | Coordinate the calcium ion. | metal ligand |
| Arg169 | Arg87A | Stabilises the transition state via bidentate hydrogen bonding and acts as an acid to protonate the 5'-hydroxyl leaving group. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, electrostatic stabiliser |
| Glu125 | Glu43A | Glu43 is acting as a general Acid/Base | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Arg117 | Arg35A | Stabilises the negatively charged transition state via bidentate hydrogen bonding. | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
proton transfer, bimolecular nucleophilic substitution, overall reactant used, overall product formed, hydrolysis, rate-determining step, native state of enzyme regenerated, inferred reaction stepReferences
- Weber DJ et al. (1992), Proteins, 13, 275-287. NMR docking of a substrate into the X-ray structure of staphylococcal nuclease. DOI:10.1002/prot.340130402. PMID:1518799.
- Libson AM et al. (1994), Biochemistry, 33, 8007-8016. Crystal structures of the binary Ca2+ and pdTp complexes and the ternary complex of the Asp21-->Glu mutant of staphylococcal nuclease. Implications for catalysis and ligand binding. DOI:10.2210/pdb1enc/pdb. PMID:8025105.
- Hale SP et al. (1993), Biochemistry, 32, 7479-7487. Mechanism of the reaction catalyzed by staphylococcal nuclease: Identification of the rate-determining step. DOI:10.1021/bi00080a020. PMID:8338846.
- Pourmotabbed T et al. (1990), Biochemistry, 29, 3677-3683. Kinetic and conformational effects of lysine substitutions for arginines 35 and 87 in the active site of staphylococcal nuclease. DOI:10.1021/bi00467a013. PMID:2111164.
- Serpersu EH et al. (1987), Biochemistry, 26, 1289-1300. Kinetic and magnetic resonance studies of active-site mutants of staphylococcal nuclease: factors contributing to catalysis. DOI:10.1021/bi00379a014. PMID:3567171.

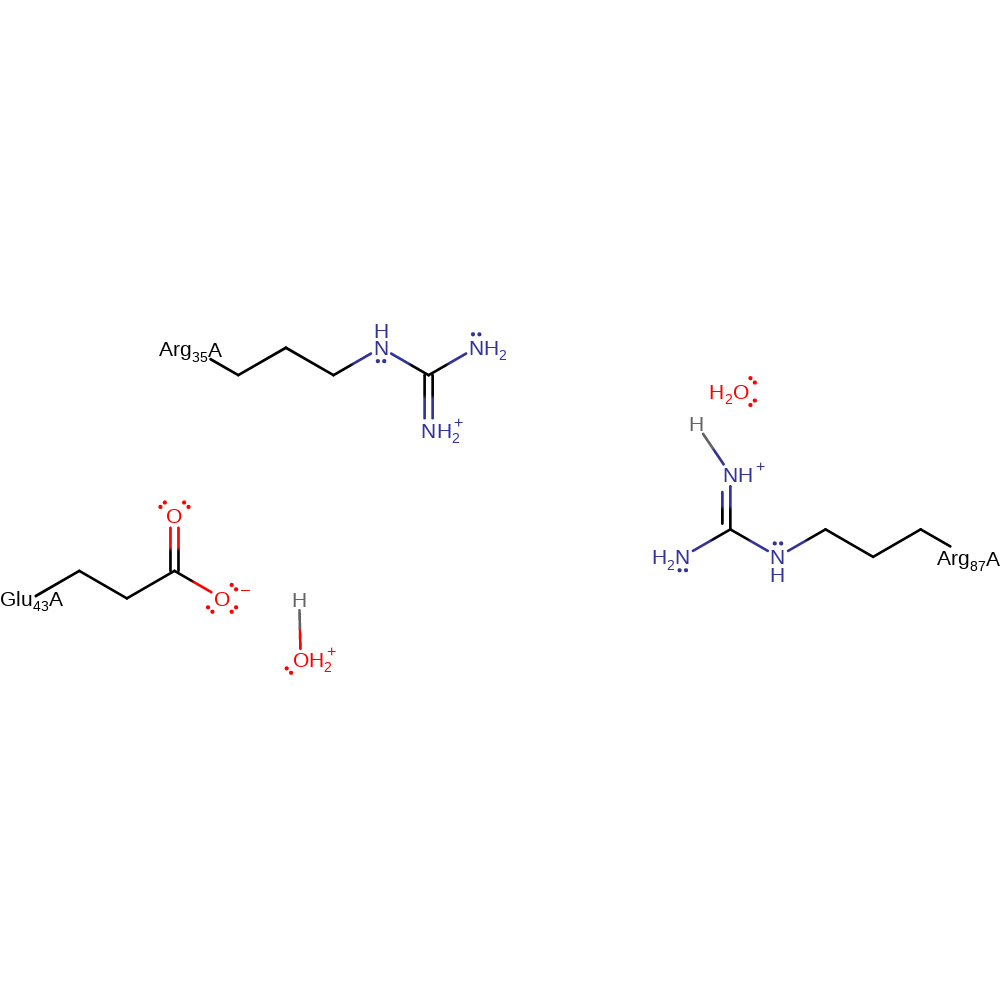
Step 1. Glu43 deprotonates the calcium activated water molecule, which then attacks the DNA phosphate bond in a nucleophilic substitution. The 3' DNA hydroxyl protonates from Arg87.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu43A | hydrogen bond acceptor |
| Arg35A | hydrogen bond donor, electrostatic stabiliser |
| Arg87A | hydrogen bond donor, electrostatic stabiliser |
| Asp21A | metal ligand |
| Asp40A | metal ligand |
| Thr41A (main-N) | metal ligand |
| Glu43A | proton acceptor |
| Arg87A | proton donor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic substitution, overall reactant used, overall product formed, hydrolysis, rate-determining step
Step 2. Arg87 deprotonates water, and water deprotonates Glu43 in an inferred step.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu43A | hydrogen bond donor |
| Arg87A | hydrogen bond acceptor |
| Glu43A | proton donor |
| Arg87A | proton acceptor |





 Download:
Download: