HIV-1 retropepsin
HIV-1 protease is an essential part of the viral replication machinery, responsible for processing translated proteins into functional enzymes. It is part of the apartyl protease family. It cleaves the amide bond of proteins between non-specific pairs of residues.
Reference Protein and Structure
- Sequence
-
P04585
 (2.7.7.-, 2.7.7.7, 2.7.7.49, 3.1.-.-, 3.1.13.2, 3.1.26.13, 3.4.23.16)
(2.7.7.-, 2.7.7.7, 2.7.7.49, 3.1.-.-, 3.1.13.2, 3.1.26.13, 3.4.23.16)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
HIV-1 M:B_HXB2R (Virus)

- PDB
-
1a30
- HIV-1 PROTEASE COMPLEXED WITH A TRIPEPTIDE INHIBITOR
(2.0 Å)



- Catalytic CATH Domains
-
2.40.70.10
 (see all for 1a30)
(see all for 1a30)
Enzyme Reaction (EC:3.4.23.16)
Enzyme Mechanism
Introduction
Equivalent aspartate residues from each chain are involved in catalysis. In the first step, one acts as a base to deprotonate the nucleophilic water molecule, while the other acts as an acid to protonate the carbonyl group and activate it. In the second step, the acid/base roles are reversed. The carbonyl group is reformed and the C-N bond of the substrate is broken.
Both aspartate residues are required for catalytic activity. One must be protonated and the other deprotonated. Dissociation of the active dimeric form results in a complete loss of activity. Note that since this is a homodimer the assignment of chains A and B is arbitrary.
Thr26/Thr26' and Gly27/Gly27' have previously been implicated in the catalytic mechanism. However, they are more likely to be involved in stabilising the active site (Thr26/Thr26') and positioning the substrate (Gly27/Gly27').
Catalytic Residues Roles
| UniProt | PDB* (1a30) | ||
| Asp513 | Asp25A | Deprotonates the nucleophilic water molecule in the first step; protonates the leaving group in the second step. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Thr514, Thr514 | Thr26A, Thr26B | Helps stabilise the negatively charged transition state. | electrostatic stabiliser, transition state stabiliser |
| Gly515 (main-N), Gly515 (main-N) | Gly27B (main-N), Gly27A (main-N) | Stabilise and hold the catalytic aspartates in position. | hydrogen bond donor, electrostatic stabiliser |
| Asp513 | Asp25B | Protonates the carbonyl in the first step; deprotonates the resulting alcohol in the second step. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
Chemical Components
proton transfer, bimolecular nucleophilic addition, overall reactant used, intermediate formation, bimolecular elimination, intermediate collapse, intermediate terminated, overall product formed, native state of enzyme regeneratedReferences
- Liu H et al. (1996), J Mol Biol, 261, 454-469. A Combined Quantum/Classical Molecular Dynamics Study of the Catalytic Mechanism of HIV Protease. DOI:10.1006/jmbi.1996.0476. PMID:8780786.
- Krzemińska A et al. (2016), J Am Chem Soc, 138, 16283-16298. Dynamic and Electrostatic Effects on the Reaction Catalyzed by HIV-1 Protease. DOI:10.1021/jacs.6b06856. PMID:27935692.
- Tie Y et al. (2004), J Mol Biol, 338, 341-352. High Resolution Crystal Structures of HIV-1 Protease with a Potent Non-peptide Inhibitor (UIC-94017) Active Against Multi-drug-resistant Clinical Strains. DOI:10.1016/j.jmb.2004.02.052. PMID:15066436.
- Piana S et al. (2002), J Mol Biol, 319, 567-583. Role of Conformational Fluctuations in the Enzymatic Reaction of HIV-1 Protease. DOI:10.1016/s0022-2836(02)00301-7. PMID:12051929.
- Mager PP (2001), Med Res Rev, 21, 348-353. The active site of HIV-1 protease. DOI:10.1002/med.1012. PMID:11410934.
- Silva AM et al. (1996), J Mol Biol, 255, 321-340. Inhibition and catalytic mechanism of HIV-1 aspartic protease. DOI:10.1006/jmbi.1996.0026. PMID:8551523.
- Blundell TL et al. (1990), Trends Biochem Sci, 15, 425-430. The 3-D structure of HIV-1 proteinase and the design of antiviral agents for the treatment of AIDS. DOI:10.1016/0968-0004(90)90280-o. PMID:2278102.

Step 1. Asp25A deprotonates water, which then attacks the carbonyl carbon of the peptide bond in a nucleophilic addition, the resulting oxyanion then deprotonates Asp25B.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp25A | hydrogen bond acceptor |
| Gly27A (main-N) | hydrogen bond donor |
| Asp25B | hydrogen bond acceptor, hydrogen bond donor |
| Gly27B (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Thr26A | transition state stabiliser |
| Thr26B | transition state stabiliser |
| Asp25B | proton donor |
| Asp25A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, overall reactant used, intermediate formation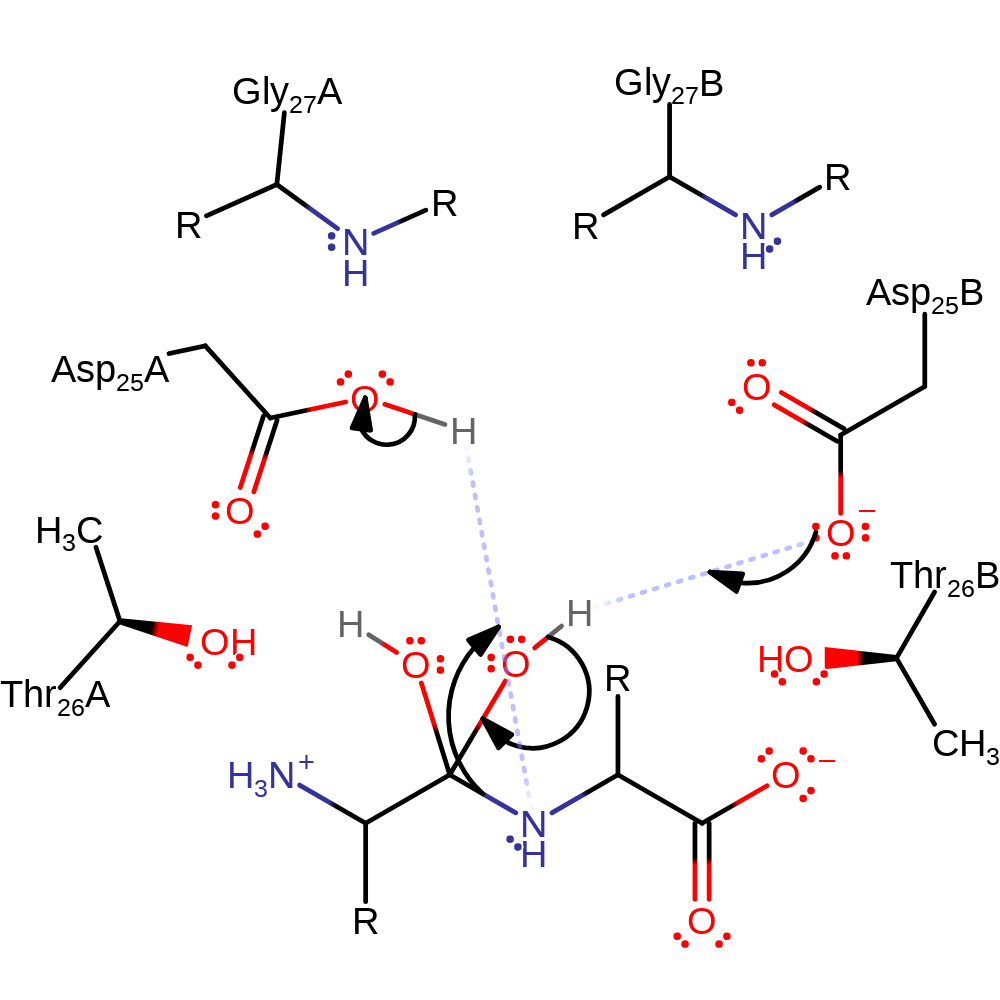
Step 2. Asp25B deprotonates the alcohol group of the intermediate, initiating an elimination that cleaves the peptide bond. The N-terminal product then deprotonates Asp25A.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr26A | electrostatic stabiliser |
| Thr26B | electrostatic stabiliser |
| Asp25A | hydrogen bond donor, hydrogen bond acceptor |
| Gly27A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Asp25B | hydrogen bond acceptor |
| Gly27B (main-N) | hydrogen bond donor |
| Asp25A | proton donor |
| Asp25B | proton acceptor |



 Download:
Download: