Thermolysin
This entry covers the metalloproteinase family M4, which contains thermolysin (EC3.4.24.27), bacillolysins (EC 3.4.24.28), pseudolysins (EC 3.4.24.26), Vibriolysin (EC3.4.24.25) and several other proteases which do not have EC numbers. Family M4 are the only members of clan MA to have structures within the PDB. The active site lies between the two domains and contains a catalytic zinc ion.
Reference Protein and Structure
- Sequence
-
P00800
 (3.4.24.27)
(3.4.24.27)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Bacillus thermoproteolyticus (Bacteria)

- PDB
-
1kei
- Thermolysin (substrate-free)
(1.6 Å)



- Catalytic CATH Domains
-
1.10.390.10
 3.10.170.10
3.10.170.10  (see all for 1kei)
(see all for 1kei)
- Cofactors
- Zinc(2+) (1) Metal MACiE
Enzyme Reaction (EC:3.4.24.27)
Enzyme Mechanism
Introduction
His231 is activated by Asp226 to act as a general acid/base. In the first step, His231 deprotonates water, which attacks the peptide bond in a nucleophilic addition, resulting in the coordination of the oxyanion to the zinc cofactor. The oxyanion collapses, eliminating the neutral terminal products. The kinetically favoured products of this reaction are the carboxyanion and the amine cation, thus in a final step the N-terminal amine deprotonates the C-terminal carboxyacid. However, there is no evidence of direct enzyme involvement in this final step [PMID:8652513].
Catalytic Residues Roles
| UniProt | PDB* (1kei) | ||
| Tyr389 | Tyr157A | Tyr157 stabilises the transition state and orients the catalytic water. | hydrogen bond donor, steric role, electrostatic stabiliser |
| Asp458 | Asp226A | Asp226 "backs up" the His231 - which acts as the general base - in a manner analogous to the Ser-His-Asp triads. | activator, hydrogen bond acceptor, electrostatic stabiliser |
| His463 | His231A | Acts as a general acid/base. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Glu375 | Glu143A | Glu143 electrostatically stabilises the cationic intermediates. | metal ligand, electrostatic stabiliser |
| His378, His374, Glu398 | His146A, His142A, Glu166A | Forms part of the zinc binding site. | metal ligand |
Chemical Components
proton transfer, bimolecular nucleophilic addition, coordination, overall reactant used, cofactor used, intermediate formation, coordination to a metal ion, rate-determining step, unimolecular elimination by the conjugate base, heterolysis, decoordination from a metal ion, intermediate collapse, native state of cofactor regenerated, native state of enzyme regenerated, intermediate terminated, overall product formedReferences
- Mock WL et al. (1996), Biochemistry, 35, 7369-7377. Arazoformyl Dipeptide Substrates for Thermolysin. Confirmation of a Reverse Protonation Catalytic Mechanism†. DOI:10.1021/bi952827p. PMID:8652513.
- Dedachi K et al. (2009), Chem Phys Lett, 479, 290-295. A combined simulation with ab initio MO and classical vibrational analysis on the specific interactions between thermolysin and dipeptide ligands. DOI:10.1016/j.cplett.2009.08.036.
- Blumberger J et al. (2007), J Chem Theory Comput, 3, 1837-1850. Peptide Hydrolysis in Thermolysin: Ab Initio QM/MM Investigation of the Glu143-Assisted Water Addition Mechanism. DOI:10.1021/ct7000792. PMID:26627626.
- Juers DH et al. (2005), Biochemistry, 44, 16524-16528. Structural Analysis of Silanediols as Transition-State-Analogue Inhibitors of the Benchmark Metalloprotease Thermolysin†,‡. DOI:10.1021/bi051346v. PMID:16342943.
- Marie-Claire C et al. (1998), FEBS Lett, 438, 215-219. Differences in transition state stabilization between thermolysin (EC 3.4.24.27) and neprilysin (EC 3.4.24.11). DOI:10.1016/s0014-5793(98)01267-8. PMID:9827548.
- Rawlings ND et al. (1995), Methods Enzymol, 248, 183-228. [13] Evolutionary families of metallopeptidases. DOI:10.1016/0076-6879(95)48015-3. PMID:7674922.

Step 1. His231 deprotonates water, which attacks the peptide bond in a nucleophilic addition, resulting in the coordination of the oxyanion to the zinc cofactor.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His142A | metal ligand |
| His146A | metal ligand |
| Glu166A | metal ligand |
| Glu143A | metal ligand |
| Glu143A | electrostatic stabiliser |
| Tyr157A | hydrogen bond donor, steric role, electrostatic stabiliser |
| His231A | hydrogen bond donor, hydrogen bond acceptor |
| Asp226A | hydrogen bond acceptor, activator |
| His231A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, coordination, overall reactant used, cofactor used, intermediate formation, coordination to a metal ion, rate-determining step
Step 2. The oxyanion initiates an elimination that cleaves the peptide bond. The N-terminal product deprotonates His231.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His142A | metal ligand |
| His146A | metal ligand |
| Glu166A | metal ligand |
| Glu143A | metal ligand |
| Glu143A | electrostatic stabiliser |
| Tyr157A | hydrogen bond donor |
| His231A | hydrogen bond donor |
| Asp226A | hydrogen bond acceptor, electrostatic stabiliser |
| His231A | proton donor |
Chemical Components
proton transfer, ingold: unimolecular elimination by the conjugate base, heterolysis, decoordination from a metal ion, intermediate collapse, intermediate formation, native state of cofactor regenerated, native state of enzyme regenerated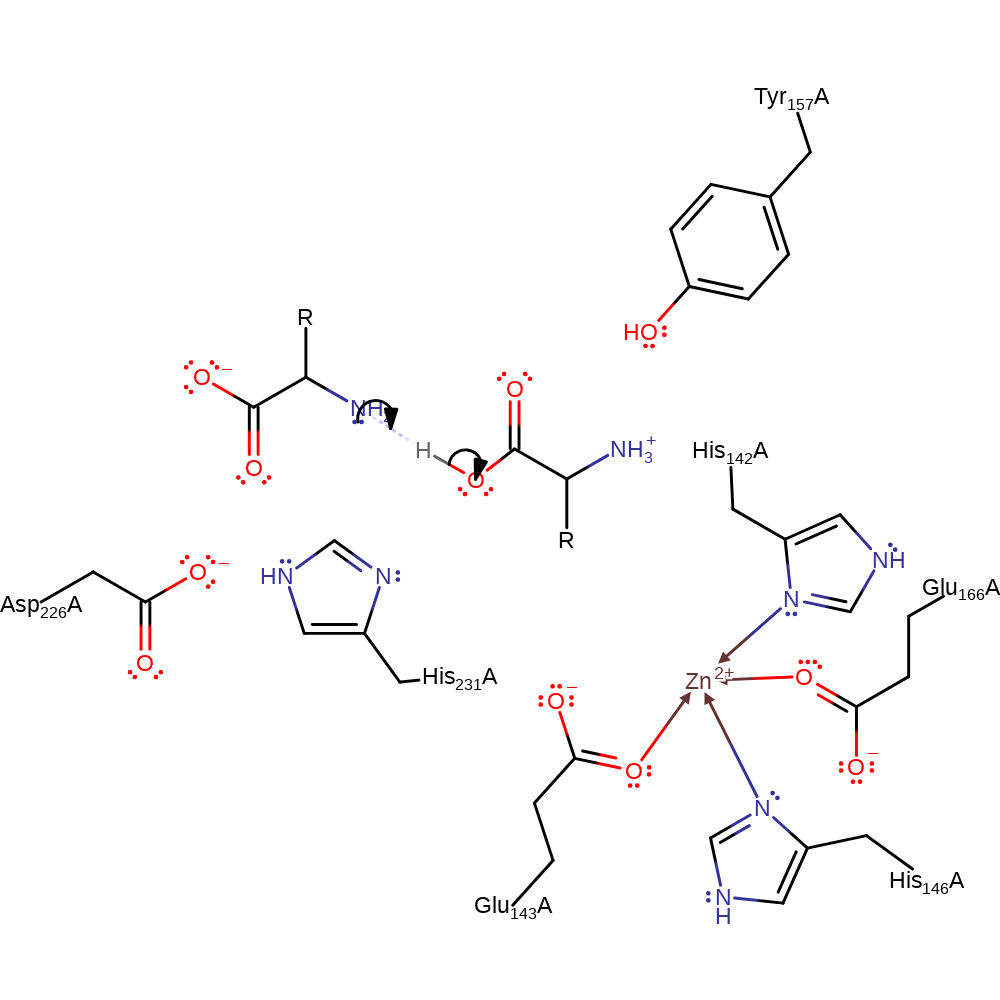
Step 3. The N-terminal product then deprotonates the C-terminal product to form the kinetically favoured products.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His142A | metal ligand |
| His146A | metal ligand |
| Glu166A | metal ligand |
| Glu143A | metal ligand |
| His231A | hydrogen bond acceptor, hydrogen bond donor |
| Asp226A | hydrogen bond acceptor |



 Download:
Download: