Deoxyribodipyrimidine photo-lyase
DNA is very sensitive to UV-radiation and other damaging agents. One result of UV radiation is the production of cyclobutane-type dimers formed from adjacent pyrimidine bases which block replication and thus have cytotoxic and mutagenic effects. Such damage is, however, effectively repaired by photolyases. Photoreactivation comprises several steps: damage recognition and binding of photolyase to DNA, photon absorption, interchromophoric energy transfer and electron transfer from the chromophore to the DNA. Two cofactors are required by the enzyme, the first, FAD, is essential for the light dependent repair process whilst the second (either 8-hydroxy-5-deazaflavin or 5,10-methenyltetrahydro-folic acid) acts as a light harvesting chromophore. The photolyases are divided into two groups according to their second cofactor.
Reference Protein and Structure
- Sequence
-
P00914
 (4.1.99.3)
(4.1.99.3)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1dnp
- STRUCTURE OF DEOXYRIBODIPYRIMIDINE PHOTOLYASE
(2.3 Å)



- Catalytic CATH Domains
-
1.10.579.10
 1.25.40.80
1.25.40.80  (see all for 1dnp)
(see all for 1dnp)
- Cofactors
- Fadh2(2-) (1), (6r)-5,10-methenyltetrahydrofolate (1)
Enzyme Reaction (EC:4.1.99.3)
Enzyme Mechanism
Introduction
The reaction can be divided into five steps. First the light-harvesting cofactor absorbs a photon then the excitation energy is transferred to the catalytic cofactor. Thirdly an electron is transferred to the pyridimine dimer in the substrate. Fourthly the C5-C5 and C6-C6 sigma bonds of the cyclobutane ring are broken and finally the electron is transferred back to the flavin and the now intact DNA dissociates from the enzyme. The inert neutral radical form of FADH. created can be reactivated by irradiation of the enzyme with white light which causes electron transfer from Trp306. This form is actually able to split the dipyridimine under 280nm light by electron transfer from Trp277.
Catalytic Residues Roles
| UniProt | PDB* (1dnp) | ||
| Asn342 | Asn341A | Helps to stabilise the reactive intermediates formed during the course of the reaction. | hydrogen bond acceptor, hydrogen bond donor, electrostatic stabiliser |
| Glu275 | Glu274A | Acts as a general acid/base. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, electrostatic stabiliser |
| Trp383, Trp278, Trp307, Trp360 | Trp382A, Trp277A, Trp306A, Trp359A | Forms part of an electron transfer chain (via the tryptophanyl radical) between FAD and the substrate. | single electron relay, single electron acceptor, single electron donor |
Chemical Components
photochemical activation, overall reactant used, cofactor used, intermediate formation, intermediate terminated, native state of cofactor regenerated, electron transfer, radical formation, proton transfer, electron relay, unimolecular homolytic elimination, radical propagation, radical termination, overall product formed, native state of enzyme regeneratedReferences
- Weber S (2005), Biochim Biophys Acta, 1707, 1-23. Light-driven enzymatic catalysis of DNA repair: a review of recent biophysical studies on photolyase. DOI:10.1016/j.bbabio.2004.02.010. PMID:15721603.
- Rousseau BJG et al. (2018), J Am Chem Soc, 140, 2853-2861. Determinants of Photolyase's DNA Repair Mechanism in Mesophiles and Extremophiles. DOI:10.1021/jacs.7b11926. PMID:29401372.
- Lee W et al. (2016), Chemistry, 22, 11371-11381. Coexistence of Different Electron-Transfer Mechanisms in the DNA Repair Process by Photolyase. DOI:10.1002/chem.201600656. PMID:27362906.
- Dreuw A et al. (2013), Phys Chem Chem Phys, 15, 19957-19969. A quantum chemical perspective on (6-4) photolesion repair by photolyases. DOI:10.1039/c3cp53313a. PMID:24145385.
- Essen LO et al. (2006), Cell Mol Life Sci, 63, 1266-1277. Light-driven DNA repair by photolyases. DOI:10.1007/s00018-005-5447-y. PMID:16699813.
- Tamada T et al. (1997), Nat Struct Biol, 4, 887-891. Crystal structure of DMA photolyase from Anacystis nidulans. DOI:10.1038/nsb1197-887. PMID:9360600.
- Park HW et al. (1995), Science, 268, 1866-1872. Crystal structure of DNA photolyase from Escherichia coli. DOI:10.1126/science.7604260. PMID:7604260.

Step 1. Photoexcitation of the 5,10-Methenyltetrahydrofolate cofactor. The function of the 5,10-Methenyltetrahydrofolate cofactor is to gather light by absorption in the regions of near-UV and visible wavelengths [PMID:15721603]. The FAD cofactor is unusually bind in the C terminal domain by adopting a U shape conformation this allows the adenine moiety to contact the redox active isoalloxazine group.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu274A | hydrogen bond acceptor, hydrogen bond donor |
| Trp277A | van der waals interaction, polar/non-polar interaction |
| Asn341A | hydrogen bond acceptor, hydrogen bond donor |
Chemical Components
photochemical activation, overall reactant used, cofactor used, intermediate formation
Step 2. The 5,10-Methenyltetrahydrofolate cofactor donates activation energy to the FAD cofactor. To achieve its active state the flavin is reduced by a light driven process to a deprotonated FADH- species utilising an electron transfer pathway involving 3 conserved tryptophan residues with the order Trp306 -> Trp359 -> Trp382 -> FAD(H.). Transfer of the excitation energy from excited MHF473 to FADH- yields the excited singlet state FADH-*. In the absence of this second chromophore (MTHF) FADH-* can also be generated by direct photoexcitation of FADH- [PMID:15721603]. Evidence suggests the transfer of energy from the antennae pigments to FADH is via a Intermolecular Coulombic Decay.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu274A | hydrogen bond acceptor, hydrogen bond donor |
| Trp277A | van der waals interaction, polar/non-polar interaction |
| Asn341A | hydrogen bond acceptor, hydrogen bond donor |
| Trp359A | single electron acceptor |
| Trp306A | single electron relay |
| Trp359A | single electron relay |
| Trp382A | single electron relay |
| Trp306A | single electron acceptor |
| Trp382A | single electron acceptor, single electron donor |
| Trp359A | single electron donor |
| Trp306A | single electron donor |
Chemical Components
photochemical activation, intermediate terminated, native state of cofactor regenerated, cofactor used, intermediate formation
Step 3. The FAD cofactor donates a single electron to Trp277, which relays it to the waiting DNA substrate. The substrate then deprotonates Glu274. Protonation of the electron-rich O4 oxygen of the 5' pyrimidine base (from the protonated Glu274) is concurrent with the electron transfer. This might be crucial to stabilise the radical anion cyclobutane pyrimidine dimer so that non-productive back-transfer of the electron onto the FADH. radical is avoided and bond cleavage is favoured [PMID:16699813]. It is thought that the electron transfer from FADH to the lesion occurs via electron tunneling.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu274A | hydrogen bond acceptor, hydrogen bond donor |
| Trp277A | van der waals interaction, polar/non-polar interaction |
| Asn341A | hydrogen bond acceptor, hydrogen bond donor, electrostatic stabiliser |
| Trp277A | electron tunneling medium |
| Glu274A | proton donor |
| Trp277A | single electron relay, single electron donor, single electron acceptor |
Chemical Components
electron transfer, radical formation, proton transfer, overall reactant used, intermediate formation, electron relay
Step 4. The radical intermediate undergoes a homolytic elimination which results in the cleavage of the four membered ring between the two nucleoside bases.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu274A | hydrogen bond acceptor, electrostatic stabiliser |
| Trp277A | van der waals interaction, polar/non-polar interaction |
| Asn341A | hydrogen bond acceptor, hydrogen bond donor, electrostatic stabiliser |
Chemical Components
ingold: unimolecular homolytic elimination, radical propagation, intermediate formation
Step 5. The radical intermediate undergoes a second homolytic elimination which results in the cleavage of the second C-C bond between the two nucleoside bases.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu274A | hydrogen bond acceptor, electrostatic stabiliser |
| Trp277A | van der waals interaction, polar/non-polar interaction |
| Asn341A | hydrogen bond acceptor, hydrogen bond donor, electrostatic stabiliser |
Chemical Components
ingold: unimolecular homolytic elimination, radical propagation, intermediate formation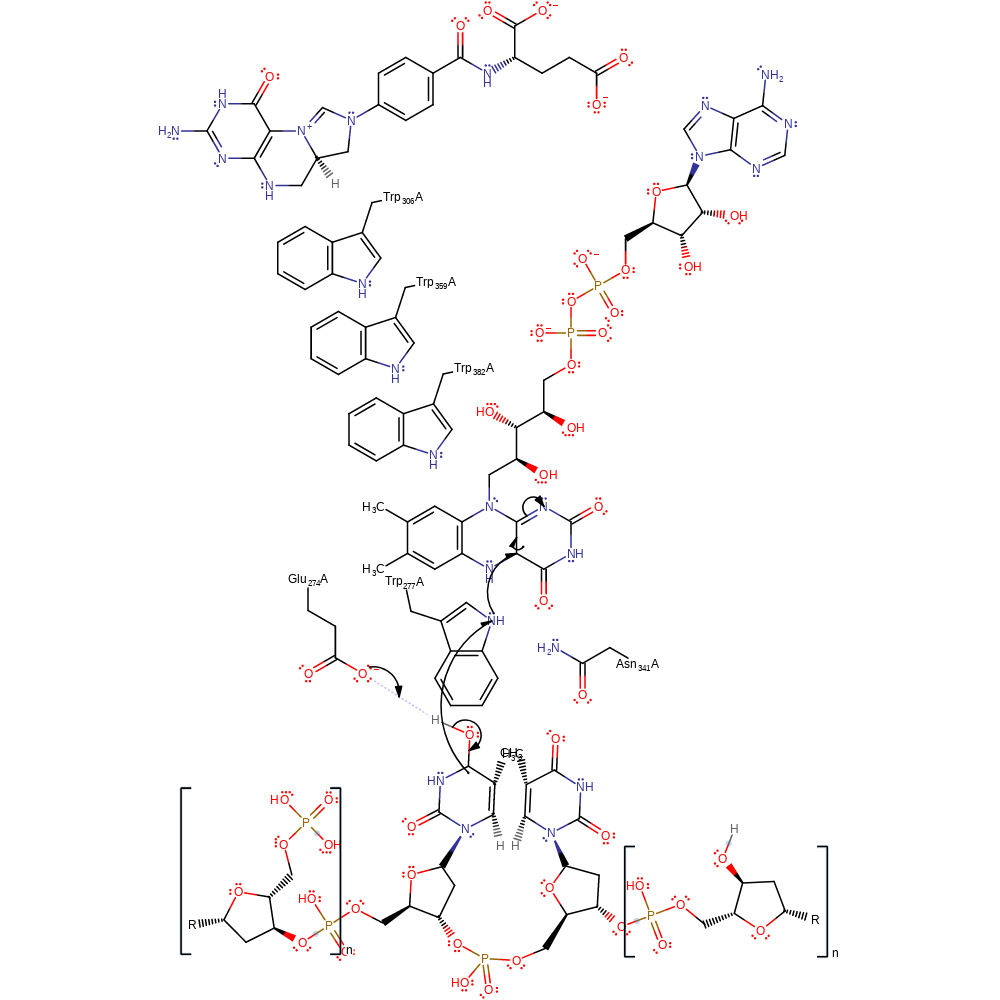
Step 6. Glu274 deprotonates the DNA radical intermediate, which results in a single electron being transferred to Trp277, which relays the electron to FAD472. Releasing the DNA product and regenerating the active site. This is again thought to involve tunneling.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu274A | hydrogen bond acceptor, electrostatic stabiliser |
| Trp277A | van der waals interaction, polar/non-polar interaction |
| Asn341A | hydrogen bond acceptor, hydrogen bond donor |
| Trp277A | electron tunneling medium |
| Glu274A | proton acceptor |
| Trp277A | single electron relay, single electron donor, single electron acceptor |



 Download:
Download: