Uroporphyrinogen-III synthase
uroporphyrinogen III synthase (URO3S) catalyses the conversion of hydroxymethylbilane (HMB) to uroporphyrinogen III, a reaction involving ring closure and intramolecular rearrangement in which the inversion of the final pyrrole unit (ring D) of the linear tetrapyrrole molecule is linked to the first pyrrole unit (ring A). Uroporphyrinogen III represents a branch point for the pathways leading to formation of a wide variety of porphyrins, including heme, chlorophyll and corrins. Porphyrins act as cofactors for a multitude of enzymes that perform a variety of processes within the cell such as methionine synthesis (vitamin B12) or oxygen transport (heme). Deficiencies of URO3S in vivo are associated with congenital erythropoietic porphyria.
Reference Protein and Structure
- Sequence
-
P10746
 (4.2.1.75)
(4.2.1.75)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Homo sapiens (Human)

- PDB
-
1jr2
- Structure of Uroporphyrinogen III Synthase
(1.84 Å)



- Catalytic CATH Domains
- (see all for 1jr2)
Enzyme Reaction (EC:4.2.1.75)
Enzyme Mechanism
Introduction
Mechanistic studies have shown that the formation of uroporphyrinogen III involves an electrophilic addition of the substrates hydroxymethyl group to C-16 that subsequently results in the cleavage of the C-15 to C-16 bond. This mechanism is referred to as the spiro-mechanism since the key intermediate is a spiropyrrolenine produced by the initial cyclisation. This mechanism is supported by the strong inhibition caused by a spiro-lactam intermediate homologue [PMID:13680202]. Modelling studies with small molecules show that the [1,5]-sigmatropic rearrangement of the substrate is unlikely, whereas fragmentation-recombination should be facile. Thus suggesting that of the various mechanisms suggested for this enzyme, the one shown in here is the most likely [DOI:10.1021/cr00105a009]. The lack of catalytic amino acid residues can be explained due to the fact that mutation of all the surface-exposed and conserved residues (mostly serine and threonine residue), and several of the other residues in the putative active site cleft, did not reveal any single residue that was absolutely required for catalysis [PMID:12196144, PMID:11689424]. This suggests that the residues may function as hydrogen bond donors and acceptors for the variety of carboxylate side chains or pyrrole nitrogens of the substrate. It is also suggested that the enzyme does not make extensive uses of charge-charge interactions with the carboxylate side chains due to the fact that only one of the conserved residues is positively charged. Instead a potentially larger number of weaker interactions may develop between ligand and polar or hydrophobic side chains. The robustness of enzyme activity to mutational analysis may indicate that no one mutation will disrupt substrate binding when a large number of other contacts are left intact [PMID:11689424]. Finally, it has been noted in modelling studies that the active site is larger than the product which suggests that a large conformational change may be required during catalysis [PMID:12196144].
Catalytic Residues Roles
| UniProt | PDB* (1jr2) |
Chemical Components
aromatic intramolecular electrophilic substitution, proton transfer, cyclisation, dehydration, intermediate formation, overall reactant used, intramolecular elimination, intermediate collapse, decyclisation, intramolecular electrophilic addition, overall product formed, intermediate terminatedReferences
- Mathews MA et al. (2001), EMBO J, 20, 5832-5839. Crystal structure of human uroporphyrinogen III synthase. DOI:10.1093/emboj/20.21.5832. PMID:11689424.
- Fortian A et al. (2011), Adv Protein Chem Struct Biol, 83, 43-74. Structural, thermodynamic, and mechanistical studies in uroporphyrinogen III synthase: molecular basis of congenital erythropoietic porphyria. DOI:10.1016/B978-0-12-381262-9.00002-1. PMID:21570665.
- Silva PJ et al. (2008), J Phys Chem B, 112, 3144-3148. Comparative density functional study of models for the reaction mechanism of uroporphyrinogen III synthase. DOI:10.1021/jp076235f. PMID:18281969.
- Frankenberg N et al. (2003), Appl Microbiol Biotechnol, 63, 115-127. Bacterial heme biosynthesis and its biotechnological application. DOI:10.1007/s00253-003-1432-2. PMID:13680202.
- Schubert HL et al. (2002), Biochem Soc Trans, 30, 595-600. Structural diversity in metal ion chelation and the structure of uroporphyrinogen III synthase. PMID:12196144.
- Battersby AR et al. (1990), Chem Rev, 90, 1261-1274. Biosynthesis of the pigments of life: mechanistic studies on the conversion of porphobilinogen to uroporphyrinogen III. DOI:10.1021/cr00105a009.

Step 1. The aromatic pi system of hydroxymethylbilane pyrrole ring D initiates an electrophilic substitution that cyclises the linear substrate and generates water.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
ingold: aromatic intramolecular electrophilic substitution, proton transfer, cyclisation, dehydration, intermediate formation, overall reactant used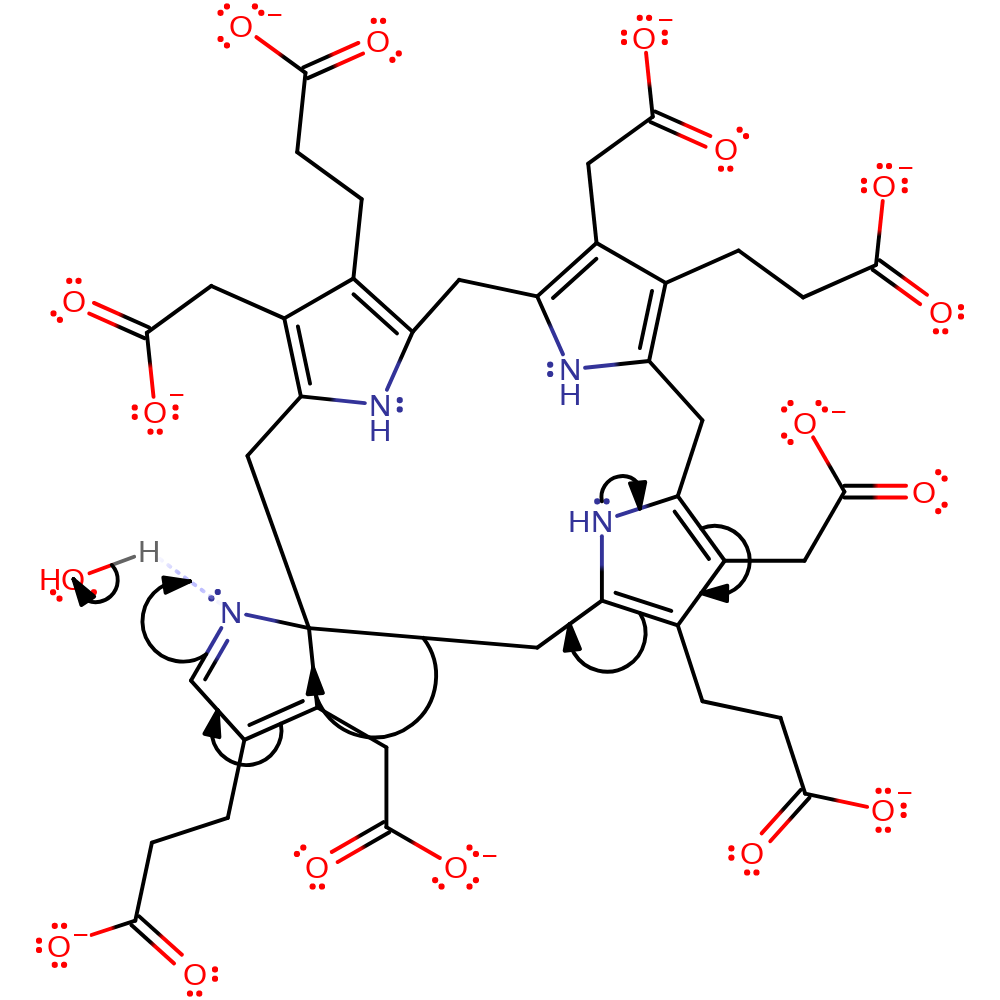
Step 2. The C pyrrole ring initiates an elimination that cleaves the C15-C16 bond between pyrrole ring D and the CH2 linker, decyclising the intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
ingold: intramolecular elimination, proton transfer, intermediate collapse, intermediate formation, decyclisation
Step 3. Pyrrole ring D initiates an electrophilic addition to the C=C group formed, re-cyclising the intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|
Chemical Components
ingold: intramolecular electrophilic addition, cyclisation, intermediate formation
Step 4. Hydroxide deprotonates pyrrole ring D, generating the final uroporphyrinogen III product.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|



 Download:
Download: