Pyruvate carboxylase
Pyruvate carboxylase, isolated from Rhizobium etli, is a biotin-dependent enzyme. It catalyses the carboxylation of pyruvate to oxaloacetate using bicarbonate and coupled to the hydrolysis of ATP to ADP and phosphate. This reaction occurs in two major steps: the carboxylation of biotin by ATP and bicarbonate in the biotin carboxylase (BC) domain, and the carboxylation of pyruvate in the carboxyltransferase (CT) domain. The biotin is bound to the biotin carboxyl carrier protein (BCCP) domain, which moves between the BC domain of one subunit and the CT domain of a neighbouring subunit.
Reference Protein and Structure
- Sequence
-
Q2K340
 (6.4.1.1)
(6.4.1.1)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Rhizobium etli CFN 42 (Bacteria)

- PDB
-
2qf7
- Crystal structure of a complete multifunctional pyruvate carboxylase from Rhizobium etli
(2.0 Å)



- Catalytic CATH Domains
-
3.20.20.70
 1.10.472.90
1.10.472.90  3.30.470.20
3.30.470.20  (see all for 2qf7)
(see all for 2qf7)
- Cofactors
- Biotinate (1), Zinc(2+) (1), Magnesium(2+) (2) Metal MACiE
Enzyme Mechanism
Introduction
BC domain: The bicarbonate is deprotonated by Glu305 and then acts as the nucleophile for attack on ATP to form ADP and phosphorylated bicarbonate. The latter undergoes spontaneous decarboxylation to form carbon dioxide and phosphate. The phosphate removes a proton from biotin to produce the biotin amidate. The carbonyl of biotin reforms and the activated biotin acts as a nucleophile through its nitrogen to attack the carbon dioxide to form carboxylbiotin.
CT domain: Pyruvate binds to the zinc ion through the oxygen of the carbonyl group. Carbon dioxide is then liberated from the biotin moiety. The activates biotin then abstracts a proton from pyruvate via Thr882. The activate substrate then initiates a nucleophilic attack on the carbon dioxide to form the final product.
Catalytic Residues Roles
| UniProt | PDB* (2qf7) | ||
| His747, His749, Asp549, Asp655 | His747(758)A, His749(760)A, Asp549(560)A, Asp655(666)A | Coordinates to the zinc ion, activating it to stabilise the formation of the pyruvate enolate. | metal ligand |
| Glu305 | Glu305(316)A | Acts as a general acid/base in the BC domain reaction. | proton acceptor, electrostatic stabiliser, proton donor |
| Arg353 | Arg353(364)A | Helps stabilise and activate the Glu305 as a general acid/base. | activator, electrostatic stabiliser |
| Thr882 | Thr882(893)A | Acts as a general acid/base in a proton relay between biotin and pyruvate in the CT domain. | proton relay, proton acceptor, proton donor |
| Glu297 | Glu297(308)A | Forms part of the binding site for both magnesium ions in the BC domain. | metal ligand |
| Glu283 | Glu283(294)A | Forms part of the binding site for magnesium 1 in the BC domain | metal ligand |
| Asn299 | Asn299(310)A | Forms part of the binding site for magnesium 2 in the BC domain. | metal ligand |
| Lys718 | Lys718(729)A | Helps stabilise the biotin intermediates. | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
proton transfer, bimolecular nucleophilic substitution, overall reactant used, intermediate formation, overall product formed, unimolecular elimination by the conjugate base, intermediate collapse, decarboxylation, assisted tautomerisation (not keto-enol), cofactor used, intermediate terminated, bimolecular nucleophilic addition, native state of enzyme is not regenerated, assisted keto-enol tautomerisation, native state of cofactor regenerated, inferred reaction step, native state of enzyme regeneratedReferences
- Menefee AL et al. (2014), FEBS J, 281, 1333-1354. Nearly 50 years in the making: defining the catalytic mechanism of the multifunctional enzyme, pyruvate carboxylase. DOI:10.1111/febs.12713. PMID:24476417.
- Lietzan AD et al. (2014), Arch Biochem Biophys, 562, 70-79. The role of biotin and oxamate in the carboxyltransferase reaction of pyruvate carboxylase. DOI:10.1016/j.abb.2014.08.008. PMID:25157442.
- Adina-Zada A et al. (2014), Biochemistry, 53, 1051-1058. Coordinating role of His216 in MgATP binding and cleavage in pyruvate carboxylase. DOI:10.1021/bi4016814. PMID:24460480.
- Lietzan AD et al. (2013), Biochem Biophys Res Commun, 441, 377-382. Insights into the carboxyltransferase reaction of pyruvate carboxylase from the structures of bound product and intermediate analogs. DOI:10.1016/j.bbrc.2013.10.066. PMID:24157795.
- Lasso G et al. (2010), Structure, 18, 1300-1310. Cryo-EM analysis reveals new insights into the mechanism of action of pyruvate carboxylase. DOI:10.1016/j.str.2010.07.008. PMID:20947019.
- Duangpan S et al. (2010), Biochemistry, 49, 3296-3304. Probing the catalytic roles of Arg548 and Gln552 in the carboxyl transferase domain of the Rhizobium etli pyruvate carboxylase by site-directed mutagenesis. DOI:10.1021/bi901894t. PMID:20230056.
- Jitrapakdee S et al. (2008), Biochem J, 413, 369-387. Structure, mechanism and regulation of pyruvate carboxylase. DOI:10.1042/bj20080709. PMID:18613815.
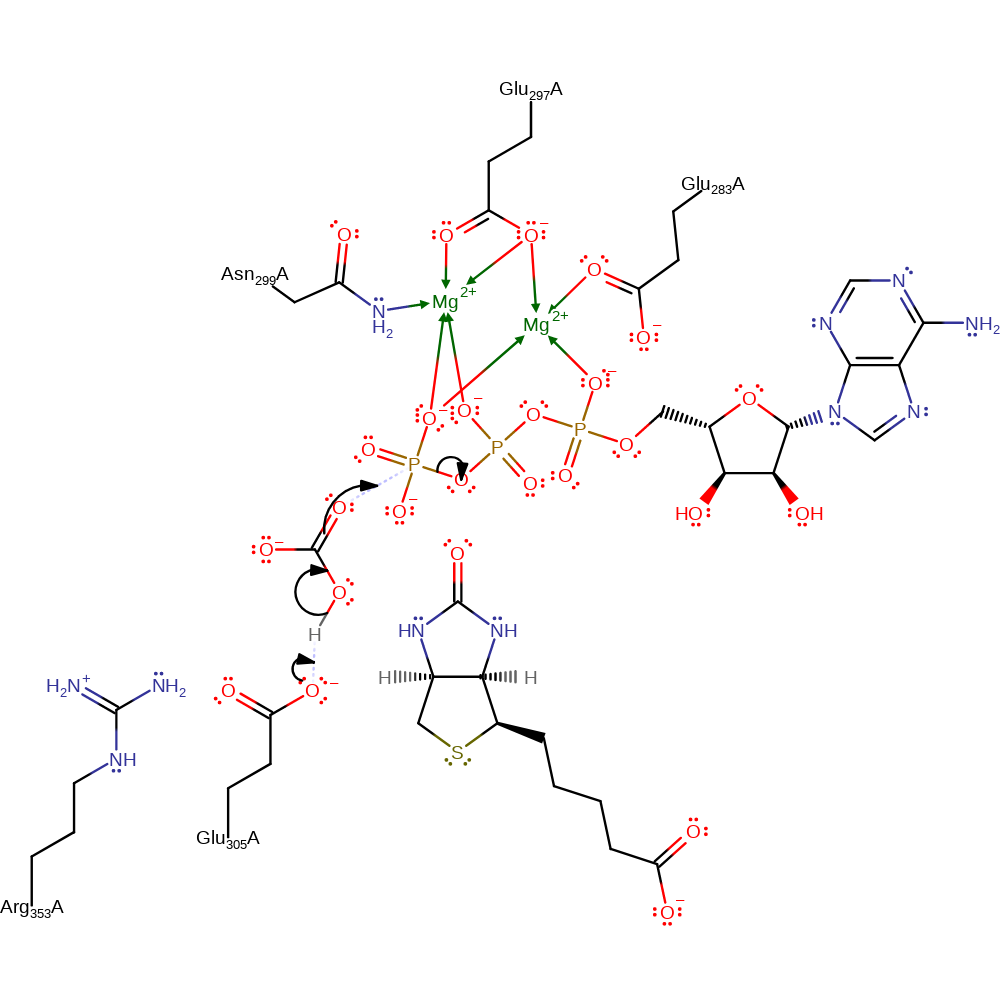
Step 1. Reaction occurs in the BC domain. The bicarbonate is deprotonated by Glu305 and then acts as a nucleophile and attacks the gamma-phosphate in a substitution reaction, liberating ADP. Mg(II) stabilises/activates the ATP
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg353(364)A | activator |
| Arg353(364)A | electrostatic stabiliser |
| Glu297(308)A | metal ligand |
| Asn299(310)A | metal ligand |
| Glu283(294)A | metal ligand |
| Glu305(316)A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic substitution, overall reactant used, intermediate formation, overall product formed
Step 2. Reaction occurs in the BC domain. The phosphorylated bicarbonate undergoes a decarboxylation reaction (E1cb) to liberate carbon dioxide and phosphate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg353(364)A | electrostatic stabiliser |
| Glu305(316)A | electrostatic stabiliser |
| Glu297(308)A | metal ligand |
| Asn299(310)A | metal ligand |
| Glu283(294)A | metal ligand |
Chemical Components
ingold: unimolecular elimination by the conjugate base, intermediate collapse, intermediate formation, decarboxylation
Step 3. Reaction occurs in the BC domain. The phosphate deprotonates one of the N-H groups of biotin with concomitant tautomerisation to produce an oxyanion.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu297(308)A | metal ligand |
| Asn299(310)A | metal ligand |
| Glu283(294)A | metal ligand |
Chemical Components
proton transfer, assisted tautomerisation (not keto-enol), cofactor used, intermediate formation, intermediate terminated, overall product formed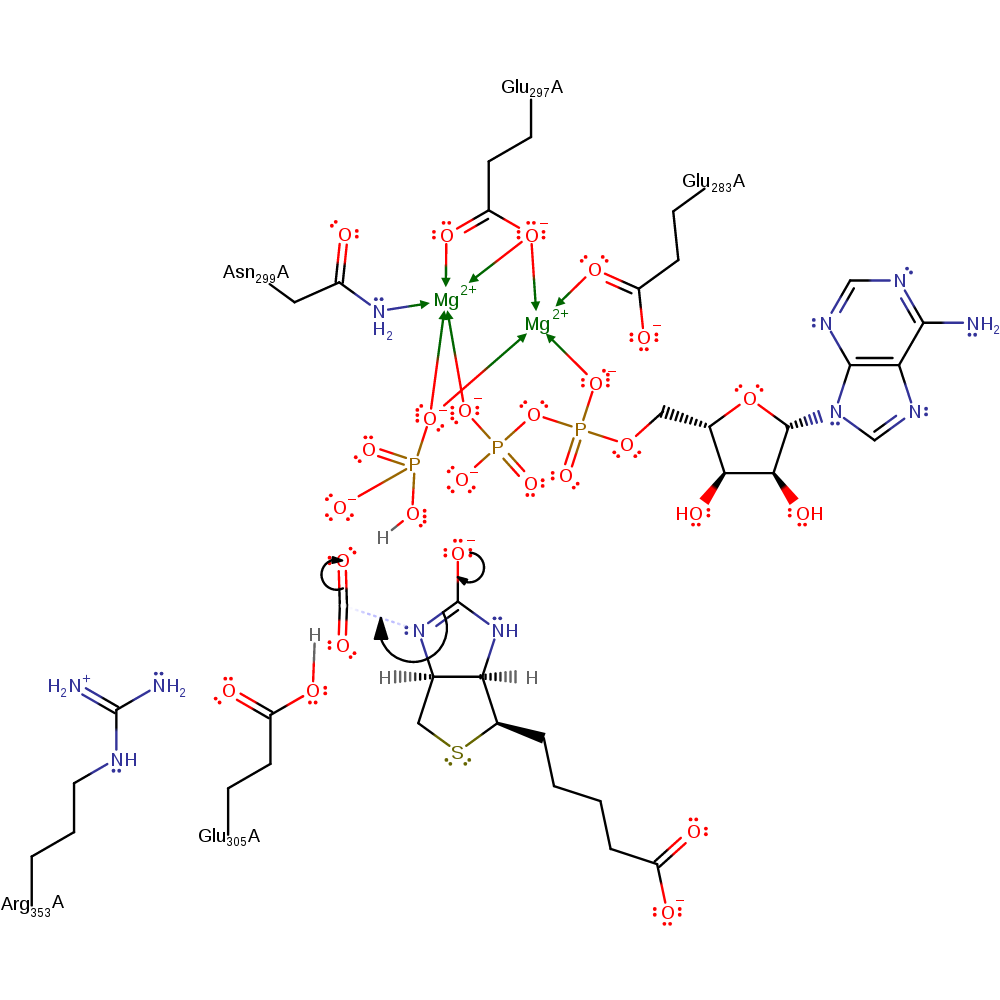
Step 4. Reaction occurs in the BC domain. The oxyanion re-forms the carbonyl group, causing the C=N bond of the activated biotin to add to the carbon dioxide in a nucleophilic manner.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg353(364)A | electrostatic stabiliser |
| Glu305(316)A | electrostatic stabiliser |
| Glu297(308)A | metal ligand |
| Asn299(310)A | metal ligand |
| Glu283(294)A | metal ligand |
Chemical Components
ingold: bimolecular nucleophilic addition, intermediate formation, native state of enzyme is not regenerated
Step 5. Reaction occurs in the CT domain. Carbon dioxide is eliminated from the biotin moiety.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys718(729)A | hydrogen bond donor, electrostatic stabiliser |
| Asp549(560)A | metal ligand |
| Asp655(666)A | metal ligand |
| His749(760)A | metal ligand |
| His747(758)A | metal ligand |
Chemical Components
intermediate formation, ingold: unimolecular elimination by the conjugate base, decarboxylation
Step 6. The activated biotin moiety abstracts a proton from Thr882, which in turn abstracts a proton from the pyruvate substrate forming the enol intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys718(729)A | electrostatic stabiliser |
| Asp549(560)A | metal ligand |
| Asp655(666)A | metal ligand |
| His749(760)A | metal ligand |
| His747(758)A | metal ligand |
| Thr882(893)A | proton donor, proton acceptor, proton relay |
Chemical Components
assisted keto-enol tautomerisation, proton transfer, native state of cofactor regenerated, overall reactant used
Step 7. The enolate intermediate initiates a nucleophilic attack on the carbon dioxide molecule to form the final product.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp549(560)A | metal ligand |
| Asp655(666)A | metal ligand |
| His749(760)A | metal ligand |
| His747(758)A | metal ligand |
Chemical Components
ingold: bimolecular nucleophilic addition, overall product formed
Step 8. Inferred return step to regenerate the active site of the BC domain.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg353(364)A | electrostatic stabiliser |
| Glu297(308)A | metal ligand |
| Glu283(294)A | metal ligand |
| Asn299(310)A | metal ligand |
| Glu305(316)A | proton donor |
Chemical Components
proton transfer, inferred reaction step, native state of enzyme regeneratedIntroduction
BC domain: The bicarbonate is deprotonated by a water molecule and then acts as the nucleophile for attack on ATP to form ADP and phosphorylated bicarbonate. The latter undergoes spontaneous decarboxylation to form carbon dioxide and phosphate. The phosphate removes a proton from biotin to produce the biotin amidate. The carbonyl of biotin reforms and the activated biotin acts as a nucleophile through its nitrogen to attack the carbon dioxide to form carboxylbiotin.
CT domain: Pyruvate binds to the zinc ion through the oxygen of the carbonyl group. Asp549 removes a proton from pyruvate to form the enolate. The enolate then attacks the carboxylate of carboxylbiotin in a nucleophilic substitution. This produces oxaloacetate and the biotin amidate, which is stabilised by accepting a proton from Lys718 to produce the isourea form of biotin. Lys718 then removes this proton and biotin returns to the ureido form by accepting a proton from Asp549.
Catalytic Residues Roles
| UniProt | PDB* (2qf7) | ||
| Asp549 | Asp549(560)A | Asp549 acts as a base to produce the enolate form of pyruvate. Later it acts as an acid to return biotin to its ureido form. | hydrogen bond acceptor, hydrogen bond donor, metal ligand, proton acceptor, proton donor |
| His747, His749, Asp655 | His747(758)A, His749(760)A, Asp655(666)A | Coordinates to the zinc ion, activating it to stabilise the formation of the pyruvate enolate. | metal ligand |
| Glu305, Arg353, Thr882 | Glu305(316)A, Arg353(364)A, Thr882(893)A | These residues have no annotated function in this mechanism proposal. | |
| Glu297 | Glu297(308)A | Forms part of the binding site for both magnesium ions in the BC domain. | metal ligand |
| Glu283 | Glu283(294)A | Forms part of the binding site for magnesium 1 in the BC domain | metal ligand |
| Asn299 | Asn299(310)A | Forms part of the binding site for magnesium 2 in the BC domain. | metal ligand |
| Lys718 | Lys718(729)A | Lys718 acts as an acid during the formation of the enol form of biotin in the PC reaction. It then acts as a base and removes this proton so that the enolate form of biotin can act as a nucleophile. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
Chemical Components
proton transfer, bimolecular nucleophilic substitution, overall reactant used, intermediate formation, overall product formed, unimolecular elimination by the conjugate base, intermediate collapse, decarboxylation, assisted tautomerisation (not keto-enol), cofactor used, intermediate terminated, bimolecular nucleophilic addition, native state of enzyme is not regenerated, assisted keto-enol tautomerisation, native state of cofactor regenerated, native state of enzyme regeneratedReferences
- Jitrapakdee S et al. (2008), Biochem J, 413, 369-387. Structure, mechanism and regulation of pyruvate carboxylase. DOI:10.1042/bj20080709. PMID:18613815.
- Menefee AL et al. (2014), FEBS J, 281, 1333-1354. Nearly 50 years in the making: defining the catalytic mechanism of the multifunctional enzyme, pyruvate carboxylase. DOI:10.1111/febs.12713. PMID:24476417.
- Zeczycki TN et al. (2009), Biochemistry, 48, 4305-4313. Insight into the carboxyl transferase domain mechanism of pyruvate carboxylase from Rhizobium etli. DOI:10.1021/bi9003759. PMID:19341298.
- Adina-Zada A et al. (2008), Int J Biochem Cell Biol, 40, 1743-1752. Insights into the mechanism and regulation of pyruvate carboxylase by characterisation of a biotin-deficient mutant of the Bacillus thermodenitrificans enzyme. DOI:10.1016/j.biocel.2008.01.001. PMID:18272421.
- St Maurice M et al. (2007), Science, 317, 1076-1079. Domain Architecture of Pyruvate Carboxylase, a Biotin-Dependent Multifunctional Enzyme. DOI:10.1126/science.1144504. PMID:17717183.
- Yong-Biao J et al. (2004), Biochemistry, 43, 5912-5920. Identification of the Catalytic Residues Involved in the Carboxyl Transfer of Pyruvate Carboxylase. DOI:10.1021/bi035783q. PMID:15134465.

Step 1. Reaction occurs in the BC domain. The bicarbonate is deprotonated by a hydroxide/water molecule and then acts as a nucleophile and attacks the gamma-phosphate in a substitution reaction, liberating ADP. Mg(II) stabilises/activates the ATP
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu297(308)A | metal ligand |
| Glu283(294)A | metal ligand |
| Asn299(310)A | metal ligand |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic substitution, overall reactant used, intermediate formation, overall product formed
Step 2. Reaction occurs in the BC domain. The phosphorylated bicarbonate undergoes a decarboxylation reaction (E1cb) to liberate carbon dioxide and phosphate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu297(308)A | metal ligand |
| Glu283(294)A | metal ligand |
| Asn299(310)A | metal ligand |
Chemical Components
ingold: unimolecular elimination by the conjugate base, intermediate collapse, intermediate formation, decarboxylation
Step 3. Reaction occurs in the BC domain. The phosphate deprotonates one of the N-H groups of biotin with concomitant tautomerisation to produce an oxyanion.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu297(308)A | metal ligand |
| Glu283(294)A | metal ligand |
| Asn299(310)A | metal ligand |
Chemical Components
proton transfer, assisted tautomerisation (not keto-enol), cofactor used, intermediate formation, intermediate terminated, overall product formed
Step 4. Reaction occurs in the BC domain. The oxyanion re-forms the carbonyl group, causing the C=N bond of the activated biotin to add to the carbon dioxide in a nucleophilic manner.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Glu297(308)A | metal ligand |
| Glu283(294)A | metal ligand |
| Asn299(310)A | metal ligand |
Chemical Components
ingold: bimolecular nucleophilic addition, intermediate formation, native state of enzyme is not regenerated
Step 5. Reaction occurs in the CT domain. The Asp549 deprotonates pyruvate, activating the pyruvate in a keto-enol tautomerisation.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys718(729)A | hydrogen bond donor |
| Asp549(560)A | hydrogen bond acceptor |
| Asp655(666)A | activator |
| Asp549(560)A | metal ligand |
| Asp655(666)A | metal ligand |
| His749(760)A | metal ligand |
| His747(758)A | metal ligand |
| Asp549(560)A | proton acceptor |
Chemical Components
proton transfer, assisted keto-enol tautomerisation, intermediate formation, overall reactant used
Step 6. Reaction occurs in the CT domain. The activated pyruvate attacks the carboxylated biotin in a nucleophilic substitution, producing the oxaloacetate product and activated biotin.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys718(729)A | hydrogen bond donor |
| Asp655(666)A | activator |
| Asp549(560)A | metal ligand |
| Asp655(666)A | metal ligand |
| His749(760)A | metal ligand |
| His747(758)A | metal ligand |
| Lys718(729)A | proton donor |
Chemical Components
ingold: bimolecular nucleophilic substitution, proton transfer, overall product formed, intermediate collapse, intermediate terminated
Step 7. Reaction occurs in the CT domain. Lys718 deprotonates the activated biotin, which in turn deprotonates Asp549, returning the enzyme to its ground state.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys718(729)A | hydrogen bond acceptor |
| Asp549(560)A | hydrogen bond donor |
| Asp549(560)A | metal ligand |
| Asp655(666)A | metal ligand |
| His749(760)A | metal ligand |
| His747(758)A | metal ligand |
| Lys718(729)A | proton acceptor |
| Asp549(560)A | proton donor |







 Download:
Download:  Download:
Download: