DNA topoisomerase (type IB)
DNA topoisomerase IB (TopIB) is an enzyme that acts to relax both negative and positive supercoils generated during transcription and DNA replication. Due to of the size of the eukaryotic chromosome, removal of these supercoils can only be accomplished locally by introducing breaks into the DNA helix. TopIB mediates DNA relaxation by creating a transient single-strand break in the DNA duplex. This transient nick allows the broken strand to rotate around its intact complement, effectively removing local supercoils. Strand nicking results from the transesterification of an active-site tyrosine (Tyr-723 in human, Tyr-274 in the viral protein) at a DNA phosphodiester bond forming a 3-phosphotyrosine covalent enzyme-DNA complex. After DNA relaxation, the covalent intermediate is reversed when the released 5-OH of the broken strand reattacks the phosphotyrosine intermediate in a second transesterification reaction. The rate of religation is normally much faster than the rate of cleavage, and this ensures that the steady-state concentration of the covalent 3-phosphotyrosyl TopIB DNA complex remains low. A variety of DNA lesions and drugs have been shown to stabilize the covalent 3-phosphotyrosyl intermediate.
Unlike most TopIBs, the enzyme isolated from the vaccinia virus exhibits specificity for cleavage at a consensus sequence: 5'-(T/C)CCTT-3' in the scissile strand with cleavage occuring after the last base. The viral TopIB is also the smallest topoisomerase known and is unusual in that it is resistant to the potent chemotherapeutic agent camptothecin (CPT). CPT is a natural product that was originally discovered because of its antitumor activity and was later demonstrated to cause the accumulation of TopIB DNA adducts in vitro and in vivo. CPTs bind the covalent 3-phosphotyrosyl intermediate and specifically block DNA religation, thus converting TopIB into a DNA-damaging agent. TopIB is the sole intramolecular target of CPT, and the cytotoxic effects of CPT poisoning are S-phase specific. During DNA replication, the replication fork is thought to collide with the trapped TopIB DNA complexes, resulting in double-strand breaks and ultimately cell death.
Reference Protein and Structure
- Sequence
-
P68698
 (5.6.2.1)
(5.6.2.1)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Vaccinia virus WR (Virus)

- PDB
-
1a41
- TYPE 1-TOPOISOMERASE CATALYTIC FRAGMENT FROM VACCINIA VIRUS
(2.3 Å)



- Catalytic CATH Domains
-
3.90.15.10
 1.20.120.380
1.20.120.380  (see all for 1a41)
(see all for 1a41)
- Cofactors
- Water (1)
Enzyme Reaction (EC:5.6.2.1)
Enzyme Mechanism
Introduction
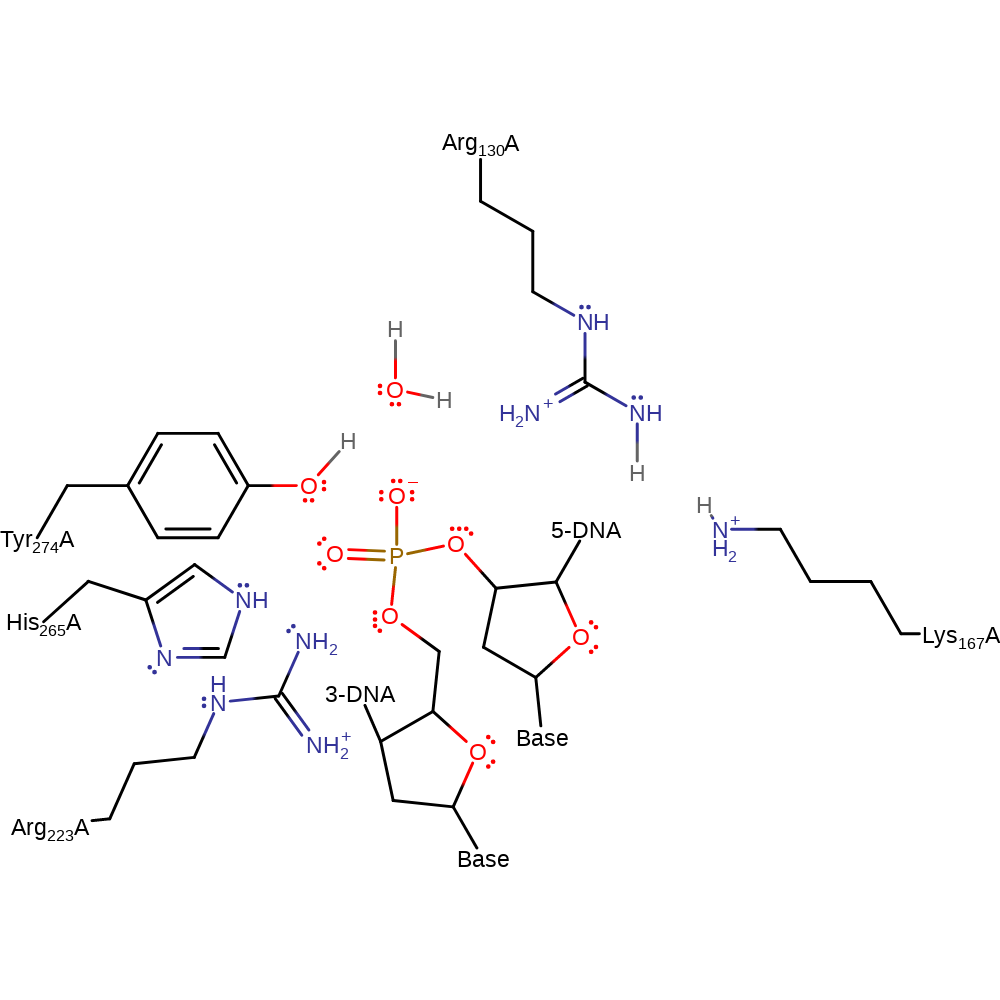
Tyr274 is the nucleophile for in-line displacement at the DNA phosphate group with the 5'-hydroxyl as the leaving group. The transition state is stabilised by Arg130 binding to one non-bridging oxygen, Arg223 and His265 binding to the other non-bridging oxygen, and Arg223 and Lys167 binding to the 5'-hydroxyl leaving group. Either Arg223 or a water molecule is involved in general base catalysis with respect to Tyr274 while Lys167 and Arg130 are involved in general acid catalysis with respect to the 5'-hydroxyl leaving group. Rotation of the DNA-enzyme intermediate around the uncleaved strand then occurs, relaxing the supercoiled DNA. The 5'-hydroxyl is then the nucleophile for in-line displacement at the DNA-enzyme intermediate. The transition state is stabilised in a similar way to that of DNA cleavage. Tyr274 is the leaving group and is protonated by Arg223/water.
Catalytic Residues Roles
| UniProt | PDB* (1a41) | ||
| Lys167, Arg130 | Lys167(87)A, Arg130(50)A | Lys167 coordinates the 5'-hydroxyl leaving group during DNA cleavage and is thought to be involved in general acid catalysis. Although the exact mechanism of protonation is uncertain, there is evidence to suggest that Arg130 and Lys167 act in a concerted fashion in the protonation of the O-5'. | attractive charge-charge interaction, hydrogen bond donor, proton acceptor, proton donor, proton relay, electrostatic stabiliser |
| Tyr274 | Tyr274(194)A | Tyr274 is the nucleophile for attack on the phosphate group during DNA cleavage. The pKa of the phenol is thought to be lowered by Arg223 to make it a better nucleophile. Arg223 may also act as a base to deprotonate Tyr274, or a water molecule could fill this role. Tyr274 is the leaving group during DNA ligation and is protonated by Arg223/water. | hydrogen bond acceptor, hydrogen bond donor, nucleophile, proton acceptor, proton donor, nucleofuge |
| Arg223 | Arg223(143)A | Arg223 stabilises the transition state of DNA cleavage by coordinating one of the phosphate non-bridging oxygens. It is though to lower the pKa of Tyr274 and may be involved in the deprotonation of Tyr274 during cleavage. If the latter is the case then it would presumably act as an acid during DNA ligation, as well as stabilising the transition state. | attractive charge-charge interaction, hydrogen bond donor, electrostatic stabiliser |
| His265 | His265(185)A | His265 stabilises the transition states of DNA cleavage and ligation by coordinating to one of the phosphate non-bridging oxygens. | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
proton transfer, bimolecular nucleophilic substitution, enzyme-substrate complex formation, intermediate formation, overall reactant used, proton relay, rate-determining step, enzyme-substrate complex cleavage, native state of enzyme regenerated, intermediate terminatedReferences
- Wittschieben J et al. (1997), Nucleic Acids Res, 25, 3001-3008. Mechanism of DNA transesterification by vaccinia topoisomerase: catalytic contributions of essential residues Arg-130, Gly-132, Tyr-136 and Lys-167. DOI:10.1093/nar/25.15.3001. PMID:9224599.
- Yakovleva L et al. (2008), J Biol Chem, 283, 16093-16103. Chemical and Traditional Mutagenesis of Vaccinia DNA Topoisomerase Provides Insights to Cleavage Site Recognition and Transesterification Chemistry. DOI:10.1074/jbc.m801595200. PMID:18367446.
- Perry K et al. (2006), Mol Cell, 23, 343-354. Structural Basis for Specificity in the Poxvirus Topoisomerase. DOI:10.1016/j.molcel.2006.06.015. PMID:16885024.
- Davies DR et al. (2006), J Mol Biol, 357, 1202-1210. The Structure of the Transition State of the Heterodimeric Topoisomerase I of Leishmania donovani as a Vanadate Complex with Nicked DNA. DOI:10.1016/j.jmb.2006.01.022. PMID:16487540.
- Interthal H et al. (2004), J Biol Chem, 279, 2984-2992. The Role of Lysine 532 in the Catalytic Mechanism of Human Topoisomerase I. DOI:10.1074/jbc.m309959200. PMID:14594810.
- Krogh BO et al. (2002), J Biol Chem, 277, 5711-5714. Proton Relay Mechanism of General Acid Catalysis by DNA Topoisomerase IB. DOI:10.1074/jbc.c100681200. PMID:11756402.
- Stivers JT et al. (2000), Biochemistry, 39, 5561-5572. Stereochemical Outcome and Kinetic Effects ofRp- andSp-Phosphorothioate Substitutions at the Cleavage Site of Vaccinia Type I DNA Topoisomerase†. DOI:10.1021/bi992429c. PMID:10820030.
- Cheng C et al. (1998), Cell, 92, 841-850. Conservation of Structure and Mechanism between Eukaryotic Topoisomerase I and Site-Specific Recombinases. DOI:10.1016/s0092-8674(00)81411-7. PMID:9529259.
- Stewart L et al. (1998), Science, 279, 1534-1541. A Model for the Mechanism of Human Topoisomerase I. DOI:10.1126/science.279.5356.1534. PMID:9488652.
- Cheng C et al. (1997), J Biol Chem, 272, 8263-8269. Mutational Analysis of 39 Residues of Vaccinia DNA Topoisomerase Identifies Lys-220, Arg-223, and Asn-228 as Important for Covalent Catalysis. DOI:10.1074/jbc.272.13.8263. PMID:9079646.
- Shuman S et al. (1989), Proc Natl Acad Sci U S A, 86, 9793-9797. Mapping the active-site tyrosine of vaccinia virus DNA topoisomerase I. DOI:10.1073/pnas.86.24.9793. PMID:2557629.

Step 1. Water deprotonates Tyr274, initiating a nucleophilic in-line substitution at the DNA phosphate with a trigonal bipyramidal transition state. The leaving 5'-hydroxyl is initially protonated by Lys167, which in turn deprotonates Arg130, which deprotonates the initial acidic water in a concerted fashion.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr274(194)A | hydrogen bond donor |
| Lys167(87)A | proton relay, hydrogen bond donor, electrostatic stabiliser, attractive charge-charge interaction |
| Arg130(50)A | hydrogen bond donor, proton relay, electrostatic stabiliser, attractive charge-charge interaction |
| His265(185)A | hydrogen bond donor, electrostatic stabiliser |
| Arg223(143)A | hydrogen bond donor, electrostatic stabiliser, attractive charge-charge interaction |
| Lys167(87)A | proton acceptor |
| Arg130(50)A | proton donor, proton acceptor |
| Tyr274(194)A | nucleophile, proton donor |
| Lys167(87)A | proton donor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic substitution, enzyme-substrate complex formation, intermediate formation, overall reactant used, proton relay, rate-determining step
Step 2. Before this catalytic step occurs the DNA-enzyme intermediate rotates around the uncleaved DNA strand, relaxing the supercoiled DNA. Water deprotonates Arg130, which in turn deprotonates Lys167, which deprotonates the rotated 5'-OH initiating a nucleophilic in-line substitution at the DNA phosphate with a trigonal bipyramidal transition state/ The eliminated Tyr274 deprotonates the initial acidic water molecule.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr274(194)A | hydrogen bond acceptor |
| Lys167(87)A | proton relay, hydrogen bond donor, electrostatic stabiliser, attractive charge-charge interaction |
| Arg130(50)A | hydrogen bond donor, proton relay, electrostatic stabiliser, attractive charge-charge interaction |
| His265(185)A | hydrogen bond donor, electrostatic stabiliser |
| Arg223(143)A | hydrogen bond donor, electrostatic stabiliser, attractive charge-charge interaction |
| Lys167(87)A | proton acceptor |
| Tyr274(194)A | proton acceptor |
| Arg130(50)A | proton donor, proton acceptor |
| Tyr274(194)A | nucleofuge |
| Lys167(87)A | proton donor |

 Download:
Download: