Tyrosine 2,3-aminomutase
Tyrosine 2,3-aminomutase catalyses the MIO-dependent deamination of L-tyrosine to form the corresponding alpha,beta-unsaturated acid (S)-beta-tyrosine. This reaction forms part of the biosynthesis of the enediyne antitumour antibiotic C-1027.
The MIO (3,5-dihydro-5-methylidene-4H-imidazol-4-one) cofactor is formed autocatalytically by cyclization and dehydration of the three amino-acid residues alanine, serine and glycine.
Reference Protein and Structure
- Sequence
-
Q8GMG0
 (4.3.1.23, 5.4.3.6)
(4.3.1.23, 5.4.3.6)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Streptomyces globisporus (Bacteria)

- PDB
-
2rjr
- Substrate mimic bound to SgTAM
(2.1 Å)



- Catalytic CATH Domains
-
1.10.275.10
 (see all for 2rjr)
(see all for 2rjr)
- Cofactors
- 2-[(1s)-1-aminoethyl]-1-carboxymethyl-5-hydroxy-4-methylimidazole (1)
Enzyme Mechanism
Introduction
This mechanism represents the amino-MIO adduct path. Here, the amine of the substrate initiates a nucleophilic attack on the 4-ene carbon of the cofactor. Tyr63 acts as a general acid/base which initiates the elimnation that results in the aminated cofactor and coumarate. Collapse of the oxyanion results in a nucleophilic attack on the coumarate intermediate in an addition reaction forming the final product and concomitant deprotonation of Tyr63.
Catalytic Residues Roles
| UniProt | PDB* (2rjr) | ||
| Tyr63 | Tyr63A | Acts as a general acid/base. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Gly70 (main-N) | Gly70A (main-N) | Helps stabilise Tyr63 in the negative state. | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
bimolecular nucleophilic addition, cofactor used, enzyme-substrate complex formation, intermediate formation, overall reactant used, bimolecular elimination, intermediate collapse, enzyme-substrate complex cleavage, unimolecular elimination by the conjugate base, native state of cofactor regenerated, native state of enzyme regenerated, intermediate terminated, overall product formedReferences
- Christianson CV et al. (2007), Biochemistry, 46, 7205-7214. The Structure ofl-Tyrosine 2,3-Aminomutase from the C-1027 Enediyne Antitumor Antibiotic Biosynthetic Pathway†,‡. DOI:10.1021/bi7003685. PMID:17516659.
- Christianson CV et al. (2007), J Am Chem Soc, 129, 15744-15745. The Mechanism of MIO-Based Aminomutases in β-Amino Acid Biosynthesis. DOI:10.1021/ja0762689. PMID:18052279.

Step 1. The amine of the substrate tyrosine initiates a nucleophilic attack upon the 4-ene carbon of the MIO cofator, resulting in double bond rearrangement and the formation of an oxyanion.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly70A (main-N) | hydrogen bond donor |
| Tyr63A | hydrogen bond acceptor |
| Gly70A (main-N) | electrostatic stabiliser |
Chemical Components
ingold: bimolecular nucleophilic addition, cofactor used, enzyme-substrate complex formation, intermediate formation, overall reactant used
Step 2. Tyr63 deprotonates the covalently bound tyrosine substrate initiating an elimination that results in the aminated cofactor and coumarate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr63A | hydrogen bond acceptor |
| Gly70A (main-N) | hydrogen bond donor |
| Gly70A (main-N) | electrostatic stabiliser |
| Tyr63A | proton acceptor |
Chemical Components
ingold: bimolecular elimination, intermediate collapse, intermediate formation, enzyme-substrate complex cleavage
Step 3. The oxyanion collapses, eliminating the ammonia group, which initiates a nucleophilic attack on the coumarate intermediate in an addition reaction with concomitant deprotonation of Tyr63
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly70A (main-N) | hydrogen bond donor |
| Tyr63A | hydrogen bond donor, hydrogen bond acceptor |
| Gly70A (main-N) | electrostatic stabiliser |
| Tyr63A | proton donor |
Chemical Components
ingold: unimolecular elimination by the conjugate base, ingold: bimolecular nucleophilic addition, native state of cofactor regenerated, enzyme-substrate complex cleavage, native state of enzyme regenerated, intermediate terminated, overall product formedIntroduction
In this alternative mechanism the aromatic ring of the substrate initiates nucleophilic attack on the 4-ene group of the cofactor, resulting in a carbocation intermediate in the aromatic ring and an oxyanion intermediate in the cofactor. Tyr63 acts as a general acid/base catalyst resulting in the carbocation being neutralized which leads to the elimination of ammonia and collapse of the substrate cofactor complex. The re-addition of ammnoia on to the substare leads to the formation of the final product along with the concomitant deprotonation of Tyr63.
Catalytic Residues Roles
| UniProt | PDB* (2rjr) | ||
| Tyr63 | Tyr63A | Acts as a general acid/base. | hydrogen bond acceptor, proton acceptor, proton donor |
| Gly70 (main-N) | Gly70A (main-N) | Helps stabilise Tyr63 in the negative state. | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
michael addition, overall reactant used, cofactor used, intermediate formation, enzyme-substrate complex formation, aromatic bimolecular electrophilic addition, proton transfer, aromatic intramolecular elimination, native state of cofactor regenerated, intermediate terminated, enzyme-substrate complex cleavage, overall product formed, bimolecular nucleophilic addition, native state of enzyme regeneratedReferences
- Christianson CV et al. (2007), J Am Chem Soc, 129, 15744-15745. The Mechanism of MIO-Based Aminomutases in β-Amino Acid Biosynthesis. DOI:10.1021/ja0762689. PMID:18052279.

Step 1. The aromatic ring of the substrate tyrosine initiates a nucleophilic attack upon the 4-ene carbon of the MIO cofator, resulting in double bond rearrangement and the formation of an oxyanion on the cofactor and the formation of a carbocation in the aromatic ring.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly70A (main-N) | electrostatic stabiliser |
| Gly70A (main-N) | hydrogen bond donor |
| Tyr63A | hydrogen bond acceptor |
Chemical Components
michael addition, overall reactant used, cofactor used, intermediate formation, enzyme-substrate complex formation, ingold: aromatic bimolecular electrophilic addition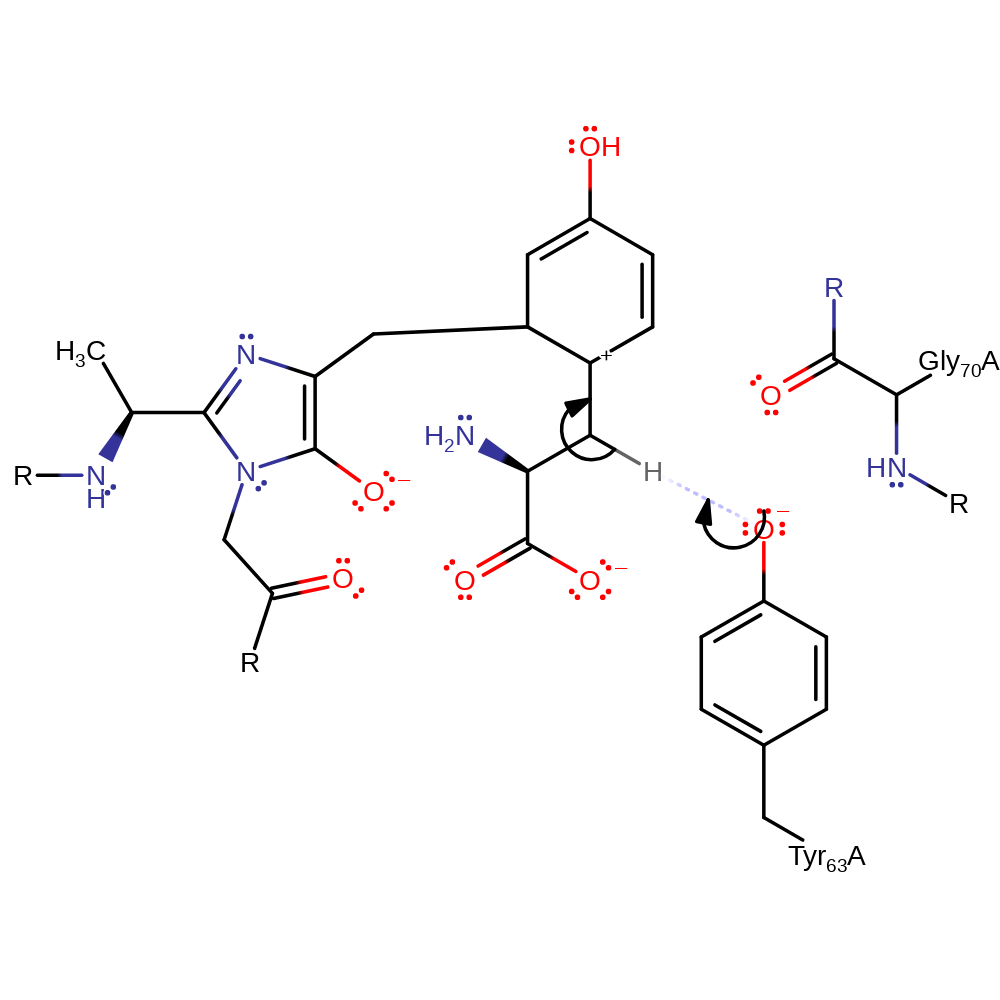
Step 2. Tyr63 deprotonates the substrate tyrosine, resulting in a C=C bond being formed and the neutralization of the carbocation.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly70A (main-N) | electrostatic stabiliser |
| Tyr63A | hydrogen bond acceptor |
| Gly70A (main-N) | hydrogen bond donor |
| Tyr63A | proton acceptor |
Chemical Components
proton transfer
Step 3. The covalent bond between the cofactor and substrate is broken leading to the rearrangement of the C=C bonds which results in ammonia being eliminated.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly70A (main-N) | electrostatic stabiliser |
Chemical Components
ingold: aromatic intramolecular elimination, native state of cofactor regenerated, intermediate terminated, enzyme-substrate complex cleavage
Step 4. The ammonia attacks the newly formed C=C double bond of the substrate in an addition reaction with concomitant deprotonation of Tyr63.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly70A (main-N) | electrostatic stabiliser, hydrogen bond donor |
| Tyr63A | hydrogen bond acceptor, proton donor |


 Download:
Download:  Download:
Download: