Chloride peroxidase (cofactor free)
Chloroperoxidase is a member of the non-haem type haloperoxidases. Haloperoxidases catalyse the halogenation of organic compounds in the presence of halide ions and peroxides such as hydrogen peroxide. They are named after the most electronegative halide they are able to oxidise. The enzyme also oxidises bromide and iodide.
This entry represents the so-called cofactor free haloperoxidases that possess a Ser-His-Asp catalytic triad and require a short aliphatic acid (e.g. ethanoic acid) for activity. They belong to the alpha/beta hydrolase superfamily.
Reference Protein and Structure
- Sequence
-
O31168
 (1.11.1.10)
(1.11.1.10)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Streptomyces aureofaciens (Bacteria)

- PDB
-
1a7u
- CHLOROPEROXIDASE T
(1.75 Å)



- Catalytic CATH Domains
-
3.40.50.1820
 (see all for 1a7u)
(see all for 1a7u)
- Cofactors
- Acetic acid (1)
Enzyme Reaction (EC:1.11.1.10)
Enzyme Mechanism
Introduction
The mechanism uses the very common Ser/His/Asp triad. Histidine deprotonates serine, which initiates a nucleophilic attack on the carboxylate carbon of the substrate. The oxyanion collapses, eliminating a water molecule with concomitant deprotonation of histidine. Histidine then deprotonates the hydrogen peroxide substrate, which initiates a nucleophilic attack on the carbonyl carbon of the enzyme-substrate intermediate. The oxyanion collapses, eliminating serine with concomitant deprotonation of histidine. A chloride ion initiates a nucleophilic attack on the peroxointermediate in a substitution reaction, regenerating the carboxylic acid and producing the hypochlorous acid intermediate. The hypochlorous acid then halogenates the alkane.
The peroxoacid is protected from hydrolysis by the hydrophobic environment of the active site as chloride, bromide or iodide enter the active site via a tunnel. The halogen then attacks the distil oxygen of the peroxoacid reforming the organic acid and hypohalous acid. Hypohalous acid may leave the active site and react with a range of substrates in solution. There is some evidence to support binding of some small hydrophobic substrates to the active site to aid specific halogenation.
The reaction catalysed by the bacterial non-heme, metal-ion- and cofactor- independent haloperoxidases is not a true peroxidase reaction, but the perhydrolysis of an ester formed between the serine residue of the catalytic triad and the organic acid. In the presence of hydrogen peroxide this ester is perhydrolysed leading to the formation of the organic peracid which can then oxidise halide ions resulting in the formation of hypohalous acid which acts as the halogenating agent.
Catalytic Residues Roles
| UniProt | PDB* (1a7u) | ||
| Met100 (main-N), Phe33 (main-N) | Met99A (main-N), Phe32A (main-N) | Forms part of the oxyanion hole to stabilise negatively charged tetrahedral transition states. | hydrogen bond donor, electrostatic stabiliser |
| Asp229 | Asp228A | Stabilises the positive charge on His257 when it is protonated. | increase basicity, hydrogen bond acceptor, increase acidity |
| His258 | His257A | Accepts a proton from Ser98 increasing its nucleophilicity. Later donates this proton to the transition state as it collapses, allowing the elimination of water. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Ser99 | Ser98A | After abstraction of a proton by His257 the oxyanion attacks the substrate carbonyl group. Productively this leads to the formation of the acyl-enzyme intermediate. Attack of the intermediate by hydrogen peroxide reforms the oxyanion which accepts a proton from the intermediate to form the product. | covalently attached, hydrogen bond acceptor, hydrogen bond donor, nucleophile, proton acceptor, proton donor, nucleofuge |
Chemical Components
proton transfer, bimolecular nucleophilic addition, enzyme-substrate complex formation, intermediate formation, overall reactant used, unimolecular elimination by the conjugate base, dehydration, overall product formed, enzyme-substrate complex cleavage, intermediate collapse, bimolecular nucleophilic substitution, native state of cofactor regenerated, native state of enzyme regenerated, intermediate terminated, substitution (not covered by the Ingold mechanisms)References
- Hofmann B et al. (1998), J Mol Biol, 279, 889-900. Structural investigation of the cofactor-free chloroperoxidases. DOI:10.1006/jmbi.1998.1802. PMID:9642069.
- van Pée KH et al. (2000), Biol Chem, 381, 1-5. Enzymatic Halogenation Catalyzed via a Catalytic Triad and by Oxidoreductases. DOI:10.1515/bc.2000.001. PMID:10722044.
- Pelletier I et al. (1995), Biochim Biophys Acta, 1250, 149-157. A catalytic triad is required by the non-heme haloperoxidases to perform halogenation. DOI:10.1016/0167-4838(95)00055-y. PMID:7632719.

Step 1. His257 deprotonates Ser98, which initiates a nucleophilic attack upon the carboxylate carbon of the carboxylic acid cofactor.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser98A | hydrogen bond donor |
| His257A | hydrogen bond acceptor, hydrogen bond donor |
| Asp228A | increase basicity, hydrogen bond acceptor |
| Phe32A (main-N) | electrostatic stabiliser, hydrogen bond donor |
| Met99A (main-N) | electrostatic stabiliser, hydrogen bond donor |
| His257A | proton acceptor |
| Ser98A | nucleophile, proton donor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, enzyme-substrate complex formation, intermediate formation, overall reactant used
Step 2. The oxyanion collapses, eliminating a water molecule with concomitant deprotonation of His257.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser98A | covalently attached |
| His257A | hydrogen bond donor |
| Asp228A | hydrogen bond acceptor, increase acidity |
| Phe32A (main-N) | electrostatic stabiliser, hydrogen bond donor |
| Met99A (main-N) | electrostatic stabiliser, hydrogen bond donor |
| His257A | proton donor |
Chemical Components
ingold: unimolecular elimination by the conjugate base, proton transfer, dehydration, overall product formed, enzyme-substrate complex cleavage, intermediate formation
Step 3. His257 deprotonates the hydrogen peroxide substrate, initiating a nucleophilic attack on the carbonyl carbon of the intermediate in an addition reaction.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser98A | covalently attached |
| His257A | hydrogen bond acceptor, hydrogen bond donor |
| Asp228A | increase basicity, hydrogen bond acceptor |
| Phe32A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Met99A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| His257A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, intermediate formation, overall reactant used, enzyme-substrate complex formation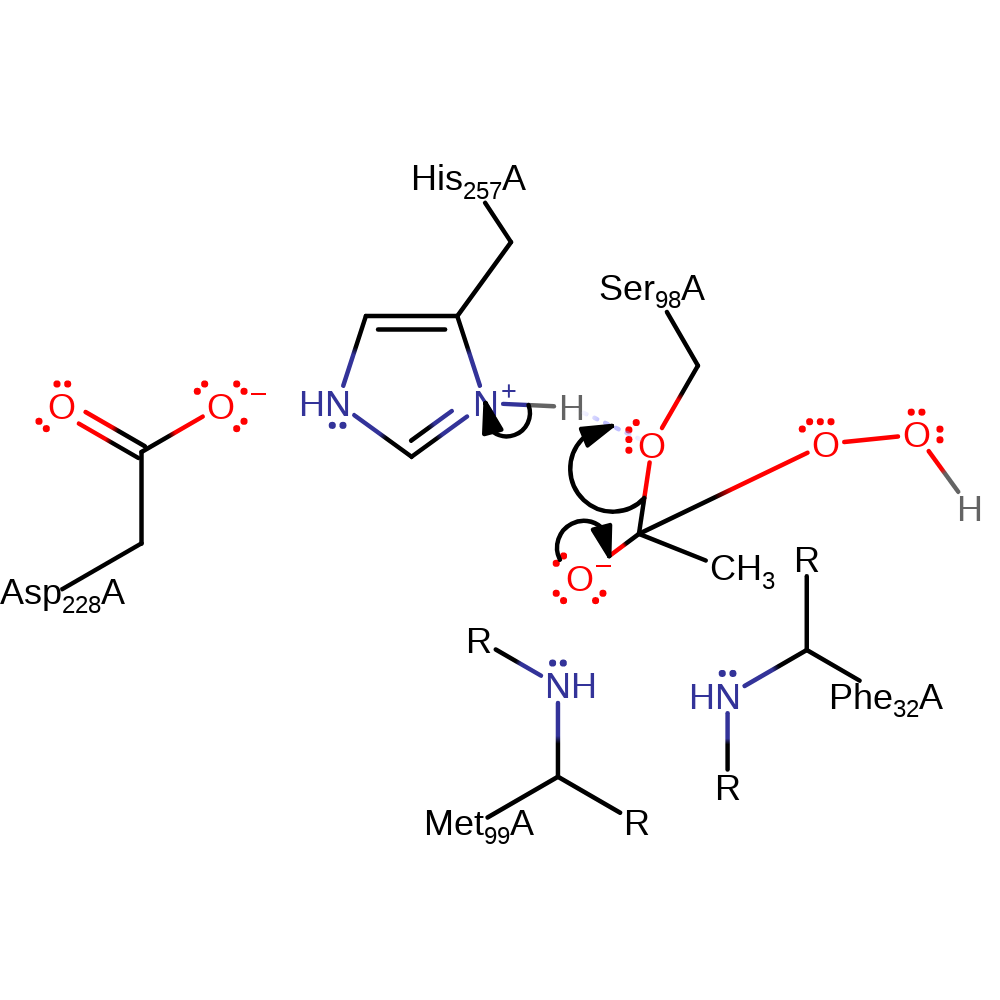
Step 4. The oxyanion collapses, eliminating the Ser98 with concomitant deprotonation of His257.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser98A | hydrogen bond acceptor |
| His257A | hydrogen bond donor |
| Asp228A | hydrogen bond acceptor, increase acidity |
| Phe32A (main-N) | electrostatic stabiliser, hydrogen bond donor |
| Met99A (main-N) | electrostatic stabiliser, hydrogen bond donor |
| Ser98A | proton acceptor, nucleofuge |
| His257A | proton donor |
Chemical Components
ingold: unimolecular elimination by the conjugate base, proton transfer, enzyme-substrate complex cleavage, intermediate collapse, intermediate formation
Step 5. A chloride ion initiates a nucleophilic attack upon the peroxo-intermediate in a substitution reaction, regenerating the carboxylic acid cofactor and producing the hypochlorous acid intermediate. Due to the fact that there does not seem to be a specific halide binding site, this step is most probably not catalysed by the enzyme. However, the hydrophobic environment of the active site pocket should protect the peroxoacid against hydrolysis whilst the halide is transported to the active site and may enhance the formation of the hypohalous acid product [PMID:9642069].
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Ser98A | hydrogen bond donor |
| His257A | hydrogen bond acceptor, hydrogen bond donor |
| Asp228A | hydrogen bond acceptor |
| Phe32A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Met99A (main-N) | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
ingold: bimolecular nucleophilic substitution, native state of cofactor regenerated, overall reactant used, native state of enzyme regenerated, intermediate formation, intermediate terminated
Step 6. The hypochlorous acid halogenates the alkane. Similarly to the previous step, the actual halogenation (which may contain multiple local minima along the reaction pathway) of the organic compound by hypohalous acid seems to be uncatalysed by the enzyme. However, it may be influenced by the size and hydrophobic environment of the active site pocket [PMID:9642069]. R represents the organic portion of the substrate to be halogenated
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp228A | hydrogen bond acceptor |






 Download:
Download: