Adenosylmethionine--8-amino-7-oxononanoate transaminase
Adenosylmethionine--8-amino-7-oxononanoate transaminase (BioA) is a PLP-dependent enzyme that functions in biotin synthesis. It utilises S-adenosyl-L-methionine (SAM) to transaminate the biotin precurson 7-keto-8-aminopelargonic acid and form the next intermediate in the pathway, 7, 8-diaminopelargonic acid. It is the only animotransferase known to utilise SAM as an amino donor. S-adenosylhomocysteine can also act as donor. SAM is a costly metabolite, however, due to the fact that the aminotransferase-catalyzed reactions are most efficient when the pKa of the substrate and the enzyme are separated, SAM may be the only substrate available with an appropriate pKa [PMID:12379100].
Reference Protein and Structure
- Sequence
-
P12995
 (2.6.1.62)
(2.6.1.62)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1dty
- CRYSTAL STRUCTURE OF ADENOSYLMETHIONINE-8-AMINO-7-OXONANOATE AMINOTRANSFERASE WITH PYRIDOXAL PHOSPHATE COFACTOR.
(2.14 Å)



- Catalytic CATH Domains
-
3.40.640.10
 3.90.1150.10
3.90.1150.10  (see all for 1dty)
(see all for 1dty)
- Cofactors
- Pyridoxal 5'-phosphate(2-) (1)
Enzyme Reaction (EC:2.6.1.62)
Enzyme Mechanism
- Summary
- Step 1
- Step 2
- Step 3
- Step 4
- Step 5
- Step 6
- Step 7
- Step 8
- Step 9
- Step 10
- Step 11
- Step 12
- Products
- All Steps
Introduction
Catalysis proceeds by a classical ping-pong bi-bi reaction mechanism. In the first steps, as in other PLP-dependent enzymes, the substrate SAM replaces Lys274 on the PLP-cofactor. Lys274 then deprotonates the CH adjacent to the bound amine, resulting in double bond rearrangement as the PLP acts as an electron sink. The PLP feeds the electrons back, resulting in the C=C attached to the aromatic ring deprotonates Lys274. Lys274 deprotonates water, which initiates a nucleophilic attack on the carbon of the C=N group in an addition reaction. The secondary amine deprotonates the attached hydroxyl group, initiating an elimination which releases S-adenosyl-4-methylthio-2-oxobutanoate. In the second phase, a Schiff base is formed between the free cofactor and 8-amino-7-oxononanoate. Lys274 then deprotonates the CH2 adjacent to the nitrogen, resulting in double bond rearrangement as the PLP acts as an electron sink. The PLP feeds the electrons back, the N+=C bond deprotonates Lys274. In the final steps of the reaction, Lys274 initiates a transaldimination resulting in the regenerated PLP cofactor and release of the 7,8-diaminononanoate poduct.
Catalytic Residues Roles
| UniProt | PDB* (1dty) | ||
| Tyr17 | Tyr17A | Only active in the second phase of the reaction. It acts to hold the second substrate in the correct orientation for the reaction to occur as well as stabilising the reactive intermediates formed during the course of the second phase of the mechanism. | hydrogen bond acceptor, hydrogen bond donor, electrostatic stabiliser, steric role |
| Tyr144 | Tyr144A | Acts to hold the free cofactor in the correct orientation to ensure that the correct reactions occur. | van der waals interaction, hydrogen bond acceptor, steric role |
| Asp245 | Asp245A | Helps stabilise the PLP cofactor to activate it as an electron sink. | increase basicity, hydrogen bond acceptor, electrostatic stabiliser, steric role |
| Lys274 | Lys274A | The main catalytic residue, in the ground state of the enzyme it is covalently attached to the PLP coafactor. During the course of the reaction it is eliminated from the cofactor, allowing it to act as a general acid/base. It acts as a nucleophile in the final transaldimination reaction. | covalently attached, hydrogen bond acceptor, hydrogen bond donor, nucleofuge, proton acceptor, proton donor, nucleophile, electron pair acceptor, electron pair donor |
Chemical Components
bimolecular nucleophilic addition, proton transfer, overall reactant used, cofactor used, enzyme-substrate complex formation, intermediate formation, unimolecular elimination by the conjugate base, enzyme-substrate complex cleavage, intermediate collapse, intramolecular elimination, overall product formed, dehydration, schiff base formed, intermediate terminated, native state of cofactor regenerated, native state of enzyme regeneratedReferences
- Eliot AC et al. (2002), Biochemistry, 41, 12582-12589. The Dual-Specific Active Site of 7,8-Diaminopelargonic Acid Synthase and the Effect of the R391A Mutation†. DOI:10.1021/bi026339a. PMID:12379100.
- Mann S et al. (2011), Biochim Biophys Acta, 1814, 1459-1466. Pyridoxal-5′-phosphate-dependent enzymes involved in biotin biosynthesis: Structure, reaction mechanism and inhibition. DOI:10.1016/j.bbapap.2010.12.004. PMID:21182990.
- Sandmark J et al. (2004), Biochemistry, 43, 1213-1222. Conserved and Nonconserved Residues in the Substrate Binding Site of 7,8-Diaminopelargonic Acid Synthase fromEscherichia coliAre Essential for Catalysis†. DOI:10.1021/bi0358059. PMID:14756557.
- Käck H et al. (1999), J Mol Biol, 291, 857-876. Crystal structure of diaminopelargonic acid synthase: evolutionary relationships between pyridoxal-5′-phosphate-dependent enzymes. DOI:10.1006/jmbi.1999.2997. PMID:10452893.

Step 1. The amine of the substrate SAM attacks the PLP cofactor in a nucleophilic addition and the bound Lys274 deprotonates the newly attached amine.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys274A | covalently attached, hydrogen bond acceptor, hydrogen bond donor |
| Asp245A | electrostatic stabiliser, hydrogen bond acceptor |
| Tyr144A | steric role, van der waals interaction |
| Lys274A | proton acceptor, electron pair acceptor |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer, overall reactant used, cofactor used, enzyme-substrate complex formation, intermediate formation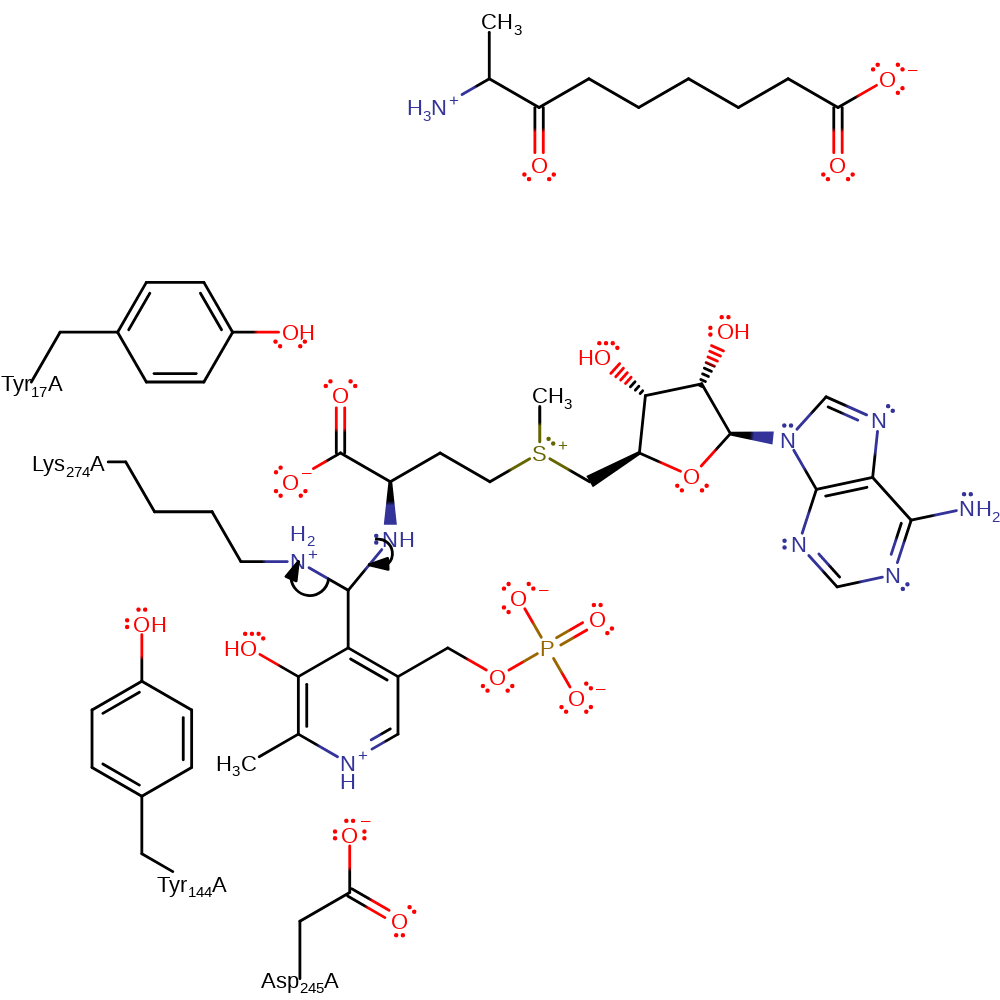
Step 2. The secondary amine that results from the initial attack initiates an elimination of the covalently bound lysine, resulting in free PLP and lysine in a neutral state.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys274A | hydrogen bond donor |
| Asp245A | electrostatic stabiliser, hydrogen bond acceptor |
| Tyr144A | steric role, van der waals interaction |
| Lys274A | nucleofuge |
Chemical Components
ingold: unimolecular elimination by the conjugate base, enzyme-substrate complex cleavage, intermediate collapse, intermediate formation
Step 3. Lys274 deprotonates the CH adjacent to the bound amine, resulting in double bond rearrangement as the PLP acts as an electron sink.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys274A | hydrogen bond acceptor, hydrogen bond donor |
| Asp245A | electrostatic stabiliser, hydrogen bond acceptor |
| Tyr144A | steric role, van der waals interaction |
| Lys274A | proton acceptor |
Chemical Components
proton transfer, intermediate formation
Step 4. The PLP feeds the electrons back, resulting in the C=C attached to the aromatic ring deprotonates Lys274.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys274A | hydrogen bond donor |
| Asp245A | hydrogen bond acceptor, increase basicity |
| Tyr144A | steric role, van der waals interaction |
| Lys274A | proton donor |
Chemical Components
proton transfer, intermediate formation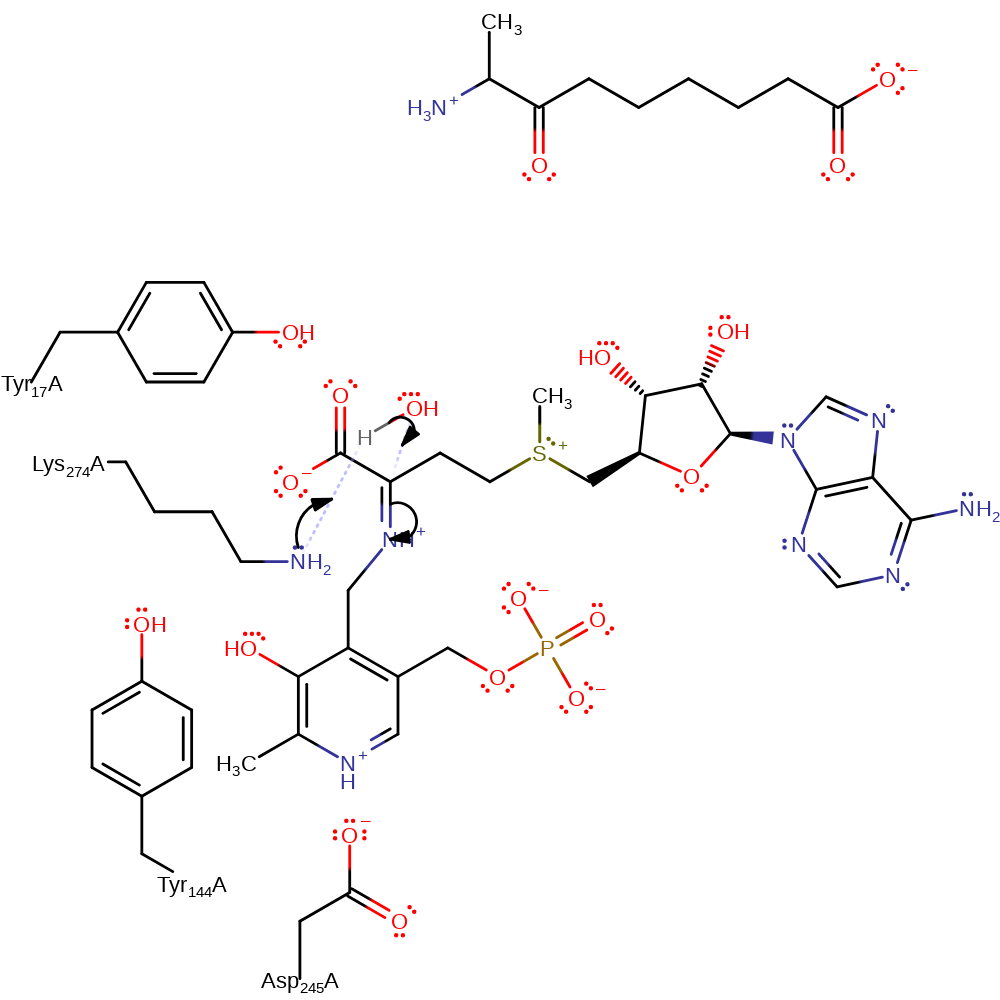
Step 5. Lys274 deprotonates water, which initiates a nucleophilic attack on the carbon of the C=N group in an addition reaction.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys274A | hydrogen bond acceptor, hydrogen bond donor |
| Asp245A | electrostatic stabiliser, hydrogen bond acceptor |
| Tyr144A | steric role, van der waals interaction |
| Lys274A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, intermediate formation
Step 6. The secondary amine deprotonates the attached hydroxyl group, initiating an elimination which releases S-adenosyl-4-methylthio-2-oxobutanoate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys274A | hydrogen bond donor |
| Asp245A | electrostatic stabiliser, hydrogen bond acceptor |
| Tyr144A | steric role, van der waals interaction |
Chemical Components
ingold: intramolecular elimination, overall product formed, intermediate collapse, intermediate formation
Step 7. The amine of PMP initiates a nucleophilic attack on the carbonyl carbon of 8-amino-7-oxononanoate. The oxyanion deprotonates the newly formed secondary amine in the first step of a Schiff base formation.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr17A | hydrogen bond donor, hydrogen bond acceptor, electrostatic stabiliser |
| Lys274A | hydrogen bond donor |
| Asp245A | steric role, hydrogen bond acceptor |
| Tyr144A | steric role, van der waals interaction, hydrogen bond acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, overall reactant used, intermediate formation
Step 8. The secondary amine initiates an elimination, forming the Schiff base and releasing water with concomitant deprotonation of Lys274.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr17A | hydrogen bond donor, hydrogen bond acceptor, steric role |
| Lys274A | hydrogen bond donor |
| Asp245A | electrostatic stabiliser, hydrogen bond acceptor |
| Tyr144A | steric role, van der waals interaction, hydrogen bond acceptor |
| Lys274A | proton donor |
Chemical Components
ingold: unimolecular elimination by the conjugate base, proton transfer, intermediate collapse, intermediate formation, dehydration, schiff base formed
Step 9. Lys274 deprotonates the CH2 adjacent to the nitrogen, resulting in double bond rearrangement as the PLP acts as an electron sink.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr17A | hydrogen bond donor, hydrogen bond acceptor, steric role |
| Lys274A | hydrogen bond acceptor, hydrogen bond donor |
| Asp245A | electrostatic stabiliser, hydrogen bond acceptor |
| Tyr144A | steric role, van der waals interaction, hydrogen bond acceptor |
| Lys274A | proton acceptor |
Chemical Components
proton transfer, intermediate formation
Step 10. The PLP feeds the electrons back, the N+=C bond deprotonates Lys274.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr17A | hydrogen bond donor, hydrogen bond acceptor, steric role |
| Lys274A | hydrogen bond donor |
| Asp245A | hydrogen bond acceptor, increase basicity |
| Tyr144A | steric role, van der waals interaction, hydrogen bond acceptor |
| Lys274A | proton donor |
Chemical Components
proton transfer, intermediate formation
Step 11. The amine of Lys274 attacks the PLP in a nucleophilic addition reaction, the secondary amine of the attached substrate reprotonates from the bound Lys274.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr17A | hydrogen bond donor, hydrogen bond acceptor, steric role |
| Lys274A | hydrogen bond donor |
| Asp245A | hydrogen bond acceptor, electrostatic stabiliser |
| Tyr144A | steric role, van der waals interaction, hydrogen bond acceptor |
| Lys274A | proton donor, nucleophile |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, enzyme-substrate complex formation, intermediate formation
Step 12. The secondary amine that results from the initial attack initiates an elimination of the covalently bound product, resulting in 7,8-diaminononanoate and the regenerated PLP cofactor.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr17A | hydrogen bond donor, steric role |
| Lys274A | covalently attached, hydrogen bond donor |
| Asp245A | hydrogen bond acceptor, electrostatic stabiliser |
| Tyr144A | steric role, van der waals interaction, hydrogen bond acceptor |
| Lys274A | electron pair donor |
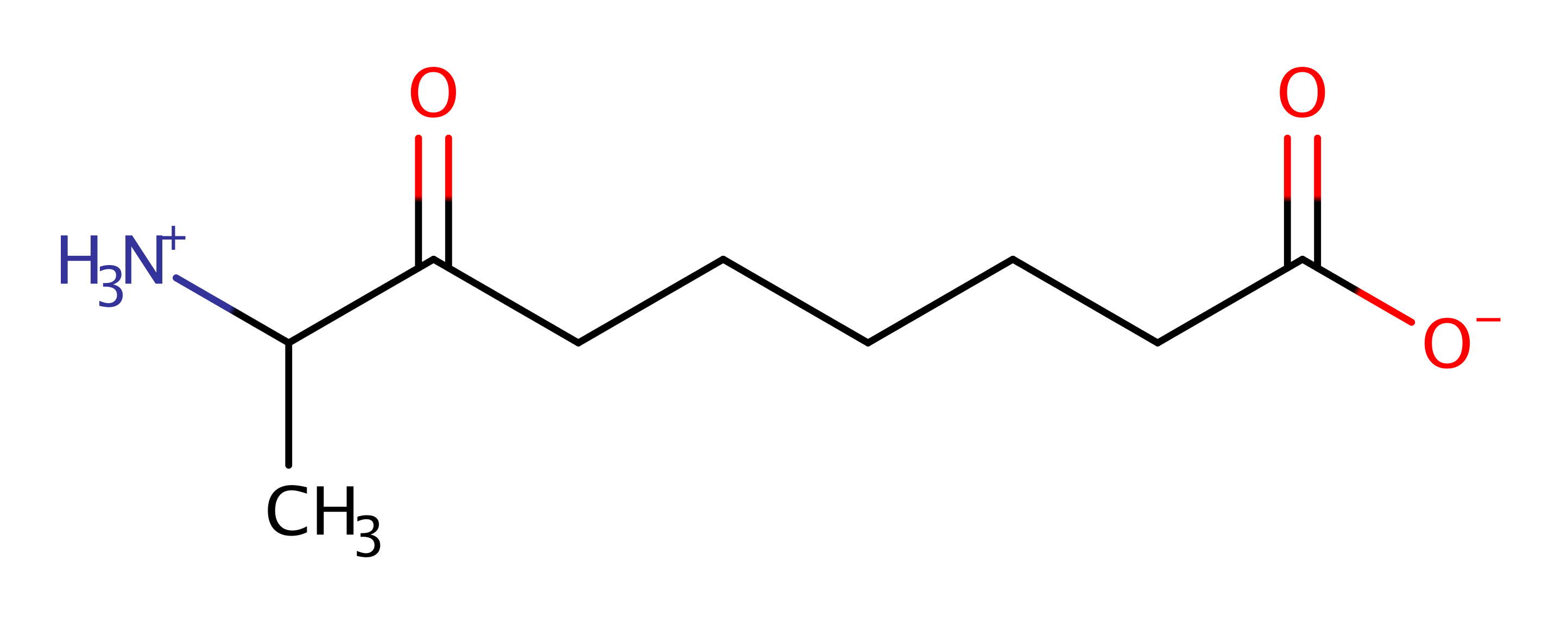



 Download:
Download: