Alcohol dehydrogenase (class I)
Alcohol dehydrogenase is the enzyme responsible for the conversion of alcohol to acetaldehyde that occurs in the cytoplasm, one of the three routes whereby acetaldehyde can be formed from alcohol before it passes into the mitochondria for the completion of the breakdown process. As a result the enzyme plays a vital role in alcohol metabolism, and inability to process alcohol is most commonly caused by deficiency.
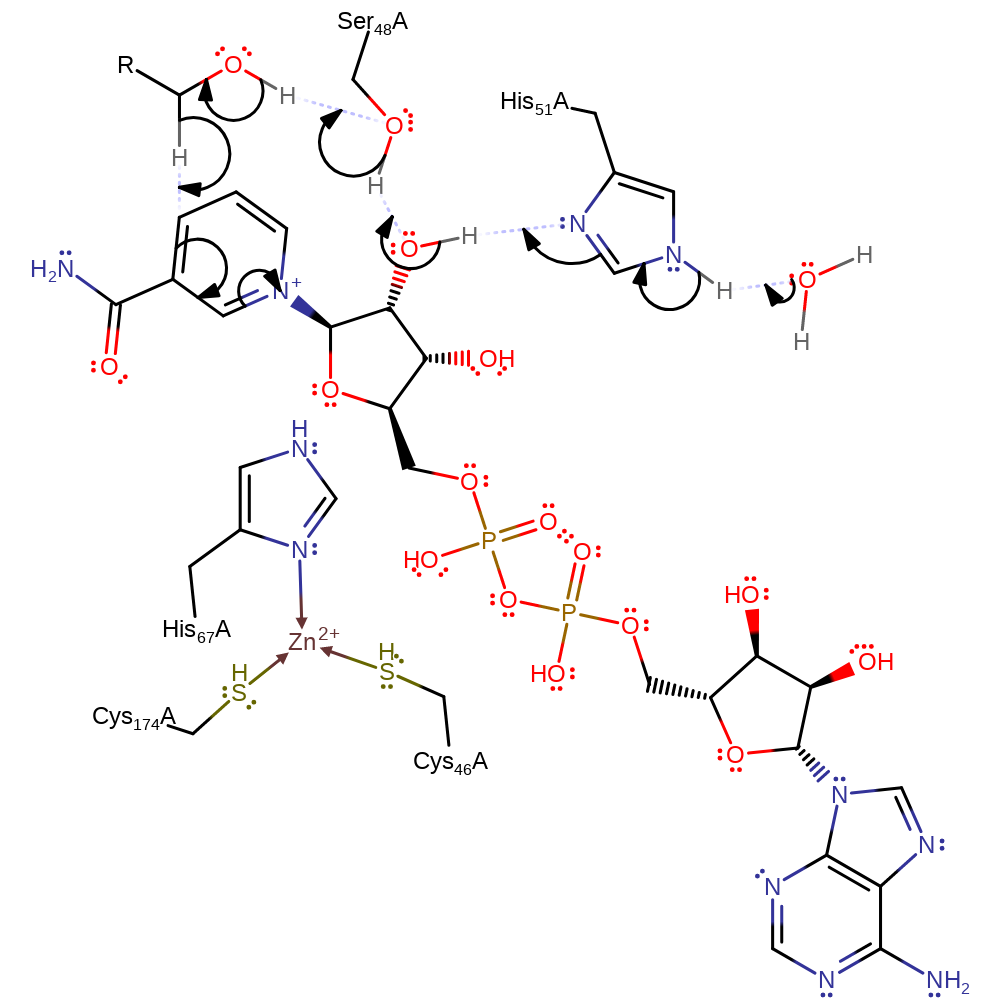
This alcohol dehydrogenase is zinc-dependent and belongs to the class I zinc-containing ADH superfamily. Whilst this enzyme binds two zinc ions, only one of these is actively involved in catalysis. It has been noted that that many ADHs that lack a His at position 51 often have an analogous histidine at position 47 which can perform the same function (PMID:10970744), i.e. activating the Ser/Thr 48 for its catalytic function, also that the residue at position 48 is interchangeable between serine and threonine (acts as a general base to deprotonate the OH group of the alcohol substrate).
Reference Protein and Structure
- Sequence
-
P00327
 (1.1.1.1)
(1.1.1.1)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Equus caballus (Horse)

- PDB
-
1qlh
- HORSE LIVER ALCOHOL DEHYDROGENASE COMPLEXED TO NAD DOUBLE MUTANT OF GLY 293 ALA AND PRO 295 THR
(2.07 Å)



- Catalytic CATH Domains
-
3.90.180.10
 (see all for 1qlh)
(see all for 1qlh)
- Cofactors
- Water (1), Nadph(4-) (1), Zinc(2+) (2) Metal MACiE
Enzyme Reaction (EC:1.1.1.1)
Enzyme Mechanism
Introduction
The enzyme is able to oxidise alcohol using concomitant reduction of NAD+. The substrate binds to a zinc ion at the active site, which allows deprotonation of the OH group by Ser 48 and His 51 in a proton relay system, because the zinc ion stabilises the oxyanion thus reducing the pKa of the OH to around 8.4. This allows hydride transfer from C2 to the NAD+ with the oxygen lone pair overlapping with the sigma* orbital of C2 to create the double bond that characterises the product.
Catalytic Residues Roles
| UniProt | PDB* (1qlh) | ||
| Ser49 | Ser48A | Acts as a general base to deprotonate the OH group of the alcohol substrate. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, proton relay |
| Cys175, Cys47, His68 | Cys174A, Cys46A, His67A | Binds catalytic zinc ion. | metal ligand |
| His52 | His51A | Reduces the pKa of Ser 48 by accepting a proton from it to allow it to act as a general base | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, proton relay |
Chemical Components
hydride transfer, proton transfer, bimolecular elimination, bimolecular nucleophilic addition, native state of enzyme regenerated, hydride relay, overall product formed, overall reactant used, proton relayReferences
- Ladenstein R et al. (2008), Cell Mol Life Sci, 65, 3918-3935. Medium- and short-chain dehydrogenase/reductase gene and protein families. DOI:10.1007/s00018-008-8590-4. PMID:19011748.
- Plapp BV (2010), Arch Biochem Biophys, 493, 3-12. Conformational changes and catalysis by alcohol dehydrogenase. DOI:10.1016/j.abb.2009.07.001. PMID:19583966.
- Hayward S et al. (2006), Biophys J, 91, 1823-1831. Molecular Dynamics Simulations of NAD+-Induced Domain Closure in Horse Liver Alcohol Dehydrogenase. DOI:10.1529/biophysj.106.085910. PMID:16714351.
- Svensson S et al. (2000), J Mol Biol, 302, 441-453. Crystal structures of mouse class II alcohol dehydrogenase reveal determinants of substrate specificity and catalytic efficiency. DOI:10.1006/jmbi.2000.4039. PMID:10970744.
- Ramaswamy S et al. (1999), Biochemistry, 38, 13951-13959. Substitutions in a Flexible Loop of Horse Liver Alcohol Dehydrogenase Hinder the Conformational Change and Unmask Hydrogen Transfer†,‡. DOI:10.1021/bi991731i. PMID:10529241.
- Al-Karadaghi S et al. (1994), Acta Crystallogr D Biol Crystallogr, 50, 793-807. Refined crystal structure of liver alcohol dehydrogenase–NADH complex at 1.8 Å resolution. DOI:10.1107/s0907444994005263. PMID:15299346.
- Eklund H et al. (1982), J Biol Chem, 257, 14349-14358. Binding of substrate in a ternary complex of horse liver alcohol dehydrogenase. PMID:6754727.




 Download:
Download: