Adenylate kinase
This family of small, monomeric enzymes is most well known for the ubiquitous Adenylate kinase (EC 2.7.4.3) which serves a crucial role in adenosine phosphate homeostasis; in times of energy need, it enables ADP to be broken down to produce some more ATP, and then when energy is once more abundant it enables the recovery of AMP produced in this and other reactions. At least two other closely related enzymes have been identified - yeast uridylate kinase (no EC no. yet) and slime mould UMP/CMP kinase (EC 2.7.4.14). In all cases Mg(2+) is required for activity.
Reference Protein and Structure
- Sequence
-
P27142
 (2.7.4.3)
(2.7.4.3)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Geobacillus stearothermophilus (Bacteria)

- PDB
-
1zio
- PHOSPHOTRANSFERASE
(1.96 Å)



- Catalytic CATH Domains
-
3.40.50.300
 (see all for 1zio)
(see all for 1zio)
- Cofactors
- Magnesium(2+) (1) Metal MACiE
Enzyme Reaction (EC:2.7.4.3)
Enzyme Mechanism
Introduction
Stereochemical studies on this enzyme indicate that the transferred phosphoryl group is inverted, indicating a single step, in-line reaction. Accordingly, structures solved with the inhibitor A-P5-A (Bis(adenosine)-5'-pentaphosphate) have enabled residues which interact with the substrate to be identified, and their position related to kinetic data. Accordingly, mutations of Lys13, Arg127, Arg160 and Arg171 (numbering for Bacillus stearothermophilus) are all found to severely hinder catalysis, and these residues are all seen to interact with the transferred phosphate, presumably to neutralise its negative charge and stabilise the transition state. In addition, mutation and NMR studies of Asp84 have demonstrated that this residue is also crucial, being primarily responsible for ligating the Mg(2+) ion.
Catalytic Residues Roles
| UniProt | PDB* (1zio) | ||
| Lys13, Arg88, Arg127, Arg160, Arg171 | Lys13A, Arg88A, Arg127A, Arg160A, Arg171A | These residues are all seen to interact with the transferred phosphate, presumably to neutralise its negative charge and stabilise the transition state. | electrostatic stabiliser |
Chemical Components
bimolecular nucleophilic substitution, overall reactant used, overall product formed, native state of enzyme regenerated, rate-determining stepReferences
- Bellinzoni M et al. (2006), Protein Sci, 15, 1489-1493. The crystal structure ofMycobacterium tuberculosisadenylate kinase in complex with two molecules of ADP and Mg2+supports an associative mechanism for phosphoryl transfer. DOI:10.1110/ps.062163406. PMID:16672241.
- Krishnamurthy H et al. (2005), Proteins, 58, 88-100. Associative mechanism for phosphoryl transfer: A molecular dynamics simulation of Escherichia coli adenylate kinase complexed with its substrates. DOI:10.1002/prot.20301. PMID:15521058.
- Berry MB et al. (1998), Proteins, 32, 276-288. Crystal structures ofBacillus stearothermophilus adenylate kinase with bound Ap5A, Mg2+ Ap5A, and Mn2+ Ap5A reveal an intermediate lid position and six coordinate octahedral geometry for bound Mg2+ and Mn2+. DOI:10.1002/(sici)1097-0134(19980815)32:3<276::aid-prot3>3.0.co;2-g. PMID:9715904.
- Ko YH et al. (1997), J Biol Chem, 272, 18875-18881. Novel Insights into the Chemical Mechanism of ATP Synthase: EVIDENCE THAT IN THE TRANSITION STATE THE -PHOSPHATE OF ATP IS NEAR THE CONSERVED ALANINE WITHIN THE P-LOOP OF THE -SUBUNIT. DOI:10.1074/jbc.272.30.18875. PMID:9228065.
- Dahnke T et al. (1992), Biochemistry, 31, 6318-6328. Mechanism of adenylate kinase. Structural and functional roles of the conserved arginine-97 and arginine-132. DOI:10.1021/bi00142a022. PMID:1627570.
- Müller CW et al. (1992), J Mol Biol, 224, 159-177. Structure of the complex between adenylate kinase from Escherichia coli and the inhibitor Ap5A refined at 1.9 A resolution. A model for a catalytic transition state. DOI:10.2210/pdb1ake/pdb. PMID:1548697.

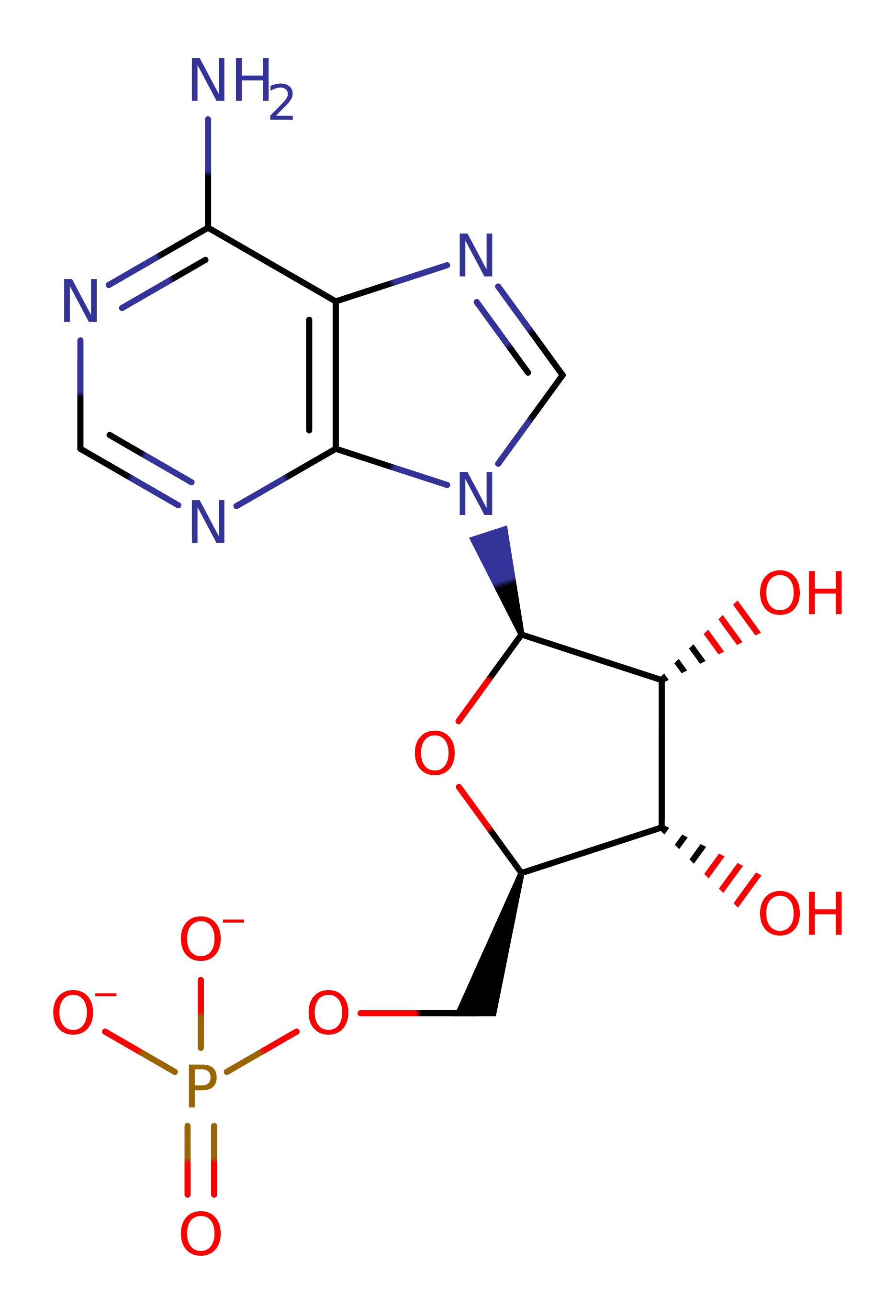
Step 1. Stereochemical studies on this enzyme suggest the transferred phosphoryl group is inverted during the course of the reaction, indicating a single step, in-line mechanism [PMID:15521058]. The magnesium divalent cation acts to neutralise the anionic ATP terminal phosphate, enhancing its electrophilicity while also directing ATP into an 'open' conformation which also facilitates the reaction [PMID:1627570, PMID:16672241].
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg127A | attractive charge-charge interaction, electrostatic stabiliser, hydrogen bond donor |
| Arg88A | attractive charge-charge interaction, electrostatic stabiliser, hydrogen bond donor |
| Arg160A | attractive charge-charge interaction, electrostatic stabiliser, hydrogen bond donor |
| Lys13A | electrostatic stabiliser |
| Arg171A | electrostatic stabiliser |


 Download:
Download: