Succinate dehydrogenase (ubiquinone)
succinate dehydrogenase is a flavoprotein complex containing iron-sulfur centres. The enzyme is found in the inner mitochondrial membrane in eukaryotes and the plasma membrane of many aerobic or facultative bacteria. It catalyses succinate oxidation in the citric acid cycle and transfers the electrons to quinones in the membrane, thus constituting a part of the aerobic respiratory chain (known as complex II). In vivo the enzyme uses the quinone found in the organism - eukaryotic enzymes utilize ubiquinone, bacterial enzymes utilise ubiquinone or menaquinone, and archaebacterial enzymes from the Sulfolobus genus use caldariellaquinone.
Reference Protein and Structure
- Sequence
-
Q9YHT1
 (1.3.5.1)
(1.3.5.1)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Gallus gallus (Chicken)

- PDB
-
1yq4
- Avian respiratory complex ii with 3-nitropropionate and ubiquinone
(2.33 Å)



- Catalytic CATH Domains
-
3.50.50.60
 3.90.700.10
3.90.700.10  (see all for 1yq4)
(see all for 1yq4)
- Cofactors
- Fadh2(2-) (1), Di-mu-sulfido-diiron(2+) (1), Tetra-mu3-sulfido-tetrairon (1), Tri-mu-sulfido-mu3-sulfido-triiron(0) (1) Metal MACiE
Enzyme Reaction (EC:1.3.5.1)
Enzyme Mechanism
Introduction
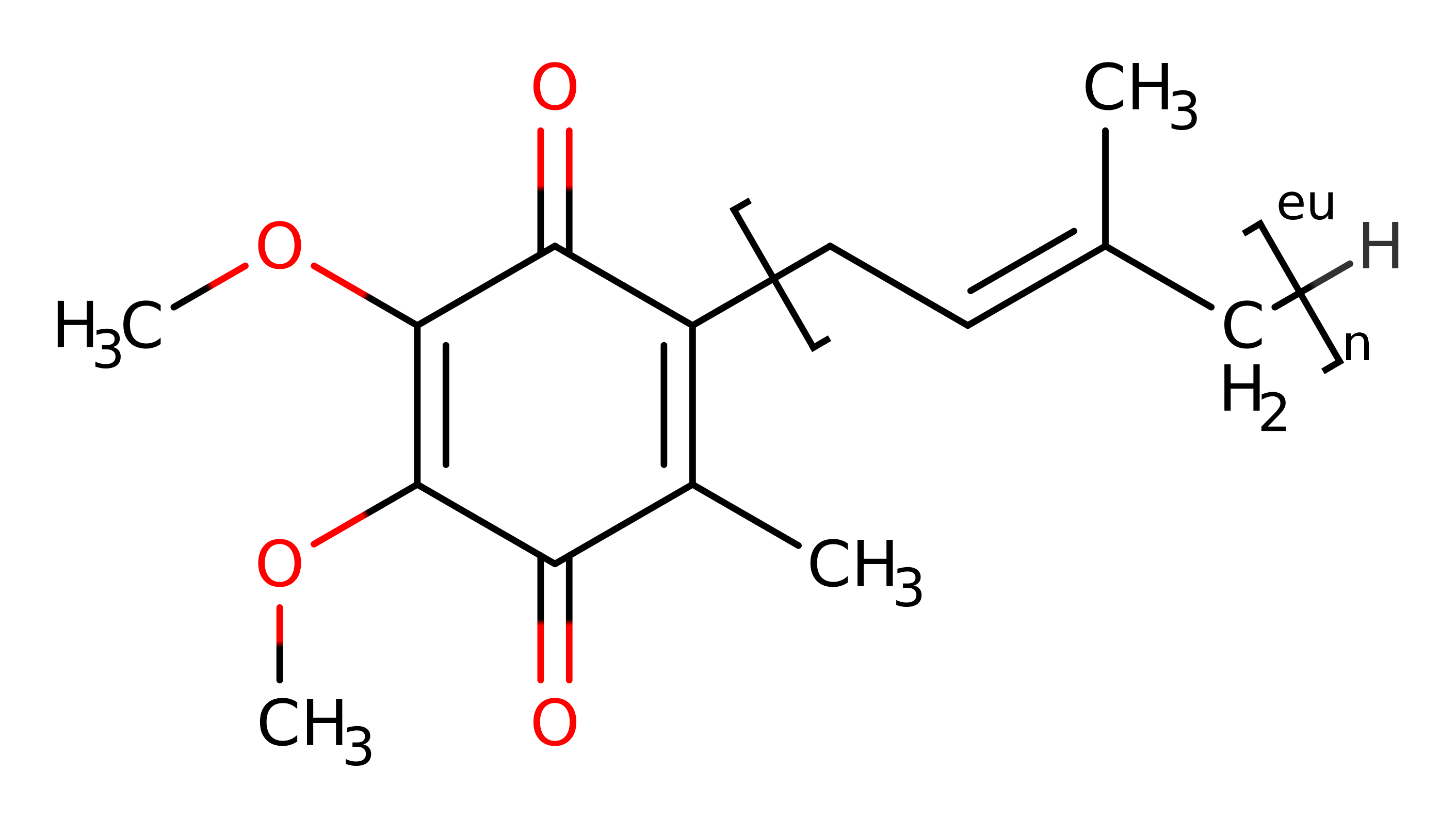
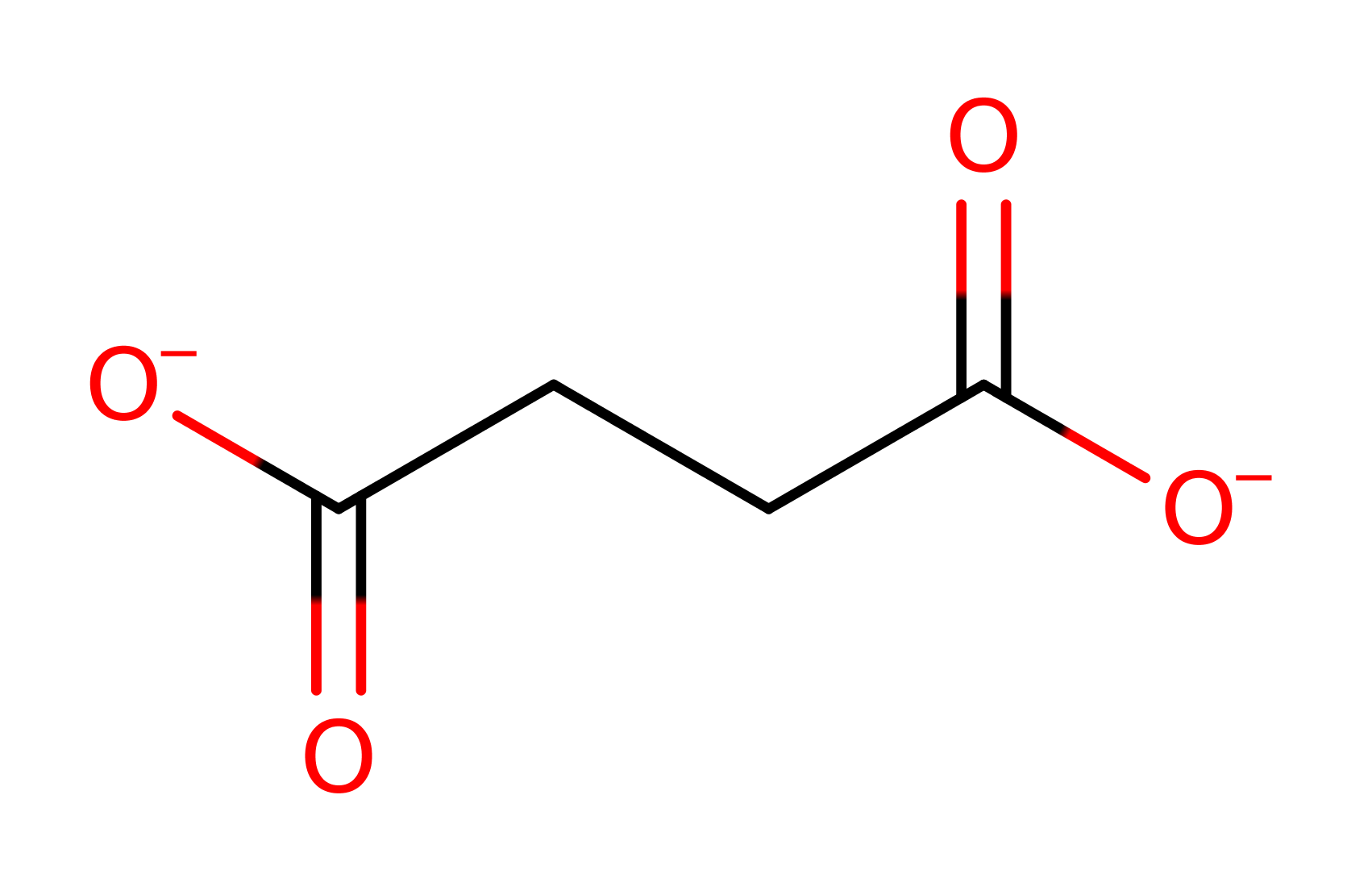
Ubiquinol acts as a two electron donor to the FAD cofactor, forming anionic FADH. The FADH N5 acts as a hydride donor to the polarised double bond in fumarate. A proton is simultaneously abstracted from Arg297 to saturate the double bond, forming succinate. The catalytic acid/base is reprotonated by a solvent molecule.
Catalytic Residues Roles
| UniProt | PDB* (1yq4) | ||
| Gln295, Glu310 | Gln251A, Glu266A | Help activate the Arg297 to act as a general acid/base. | hydrogen bond acceptor, electrostatic stabiliser, steric role |
| Arg341 | Arg297A | Acts as a general acid/base. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| His297, His408, Arg452, Leu307, Phe174 | His253A, His364A, Arg408A, Leu263A, Phe130A | The presence of residues which hydrogen bond to the two carboxylate groups of fumarate, and of bulky, non-polar residues imposes a twisted conformation within the substrate. This increases the ground state energy towards that of the transition state, while also aligning the double bond between the hydride donating FAD and proton donating Arg297 | hydrogen bond donor, steric role, electrostatic stabiliser |
Chemical Components
proton transfer, electron transfer, cofactor used, overall reactant used, overall product formed, inferred reaction step, hydride transfer, bimolecular nucleophilic addition, unimolecular elimination by the conjugate base, bond polarisation, rate-determining step, native state of enzyme regeneratedReferences
- Huang LS et al. (2006), J Biol Chem, 281, 5965-5972. 3-Nitropropionic Acid Is a Suicide Inhibitor of Mitochondrial Respiration That, upon Oxidation by Complex II, Forms a Covalent Adduct with a Catalytic Base Arginine in the Active Site of the Enzyme. DOI:10.1074/jbc.m511270200. PMID:16371358.
- Oyedotun KS et al. (2007), Biochim Biophys Acta, 1767, 1436-1445. The Saccharomyces cerevisiae succinate dehydrogenase does not require heme for ubiquinone reduction. DOI:10.1016/j.bbabio.2007.09.008. PMID:18028869.
- Zhang J et al. (2006), Proc Natl Acad Sci U S A, 103, 16212-16217. Structure of electron transfer flavoprotein-ubiquinone oxidoreductase and electron transfer to the mitochondrial ubiquinone pool. DOI:10.1073/pnas.0604567103. PMID:17050691.
- Horsefield R et al. (2006), J Biol Chem, 281, 7309-7316. Structural and Computational Analysis of the Quinone-binding Site of Complex II (Succinate-Ubiquinone Oxidoreductase). DOI:10.1074/jbc.m508173200. PMID:16407191.
- Mowat CG et al. (2001), Biochemistry, 40, 12292-12298. Kinetic and Crystallographic Analysis of the Key Active Site Acid/Base Arginine in a Soluble Fumarate Reductase†. DOI:10.1021/bi011360h. PMID:11591148.
- Taylor P et al. (1999), Nat Struct Biol, 6, 1108-1112. Structural and mechanistic mapping of a unique fumarate reductase. DOI:10.1038/70045. PMID:10581550.

Step 1.
Ubiquinol acts as a two electron donor to the FAD cofactor, forming anionic FADH.
The electrons donated by the biological reducing agent ubiquinol tunnel from the ubiquinol binding site, through the iron-sulfur clusters to the active site [PMI:16371358]. In the absence of ubiquinol, the 3Fe4S centre is left with a net positive charge [PMID:17050691].
For the related enzyme fumarate reductase, studies in E. Coli have identified residues which are thought to enhance the redox activity of ubiquinone by hydrogen bonding to the C1 carbonyl. group While they have not been shown to act as bases, the residues are conserved, including within the gallus gallus succinate dehydrogenase described here, as Trp157B and Ser39C, and could therefore be involved in activating the ubiquinol co-substrate [PMID:16407191].
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His364A | hydrogen bond donor |
| His253A | hydrogen bond donor |
| Gln251A | hydrogen bond acceptor |
| Arg297A | hydrogen bond donor |
| Arg408A | hydrogen bond donor |
| Glu266A | hydrogen bond acceptor |
Chemical Components
proton transfer, electron transfer, cofactor used, overall reactant used, overall product formed, inferred reaction step
Step 2. The FADH N5 acts as a hydride donor to the polarised double bond in fumarate. A proton is simultaneously abstracted from Arg297 to saturate the double bond, forming succinate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His364A | hydrogen bond donor, steric role, electrostatic stabiliser |
| His253A | hydrogen bond donor, steric role, electrostatic stabiliser |
| Gln251A | electrostatic stabiliser, hydrogen bond acceptor, steric role |
| Arg297A | hydrogen bond donor |
| Arg408A | hydrogen bond donor, electrostatic stabiliser, steric role |
| Leu263A | steric role |
| Glu266A | electrostatic stabiliser, hydrogen bond acceptor, steric role |
| Phe130A | steric role |
| Arg297A | proton donor |
Chemical Components
hydride transfer, proton transfer, ingold: bimolecular nucleophilic addition, ingold: unimolecular elimination by the conjugate base, bond polarisation, cofactor used, overall product formed, overall reactant used, rate-determining stepCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His364A | hydrogen bond donor |
| His253A | hydrogen bond donor |
| Gln251A | hydrogen bond acceptor |
| Arg297A | hydrogen bond donor, hydrogen bond acceptor |
| Arg408A | hydrogen bond donor |
| Glu266A | hydrogen bond acceptor |
| Arg297A | proton acceptor |



 Download:
Download: