Glycerol-3-phosphate cytidylyltransferase
Glycerol-3-phosphate cytidylyltransferase catalyses the transfer of the cytidylyl group of CTP to sn-glycerol 3-phosphate so the activated glycerol 3-phosphate can be used for the biosynthesis of teichoic acid linkage units in bacterial cell walls.
Reference Protein and Structure
- Sequence
-
P27623
 (2.7.7.39)
(2.7.7.39)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Bacillus subtilis subsp. subtilis str. 168 (Bacteria)

- PDB
-
1n1d
- Glycerol-3-phosphate cytidylyltransferase complexed with CDP-glycerol
(2.0 Å)



- Catalytic CATH Domains
-
3.40.50.620
 (see all for 1n1d)
(see all for 1n1d)
- Cofactors
- Magnesium(2+) (1) Metal MACiE
Enzyme Reaction (EC:2.7.7.39)
Enzyme Mechanism
Introduction
Proceeds via a random order reaction mechanism where there is negative cooperativity in the binding of substrates but not in catalysis. Several residues has been implicated in regulating a negative cooperativity between the binding of glycerol-3-phosphate and CTP. However, the NMR and crystallographic data characterising such residues are inconsistent, and as of yet have not explicitly identified the role that individual residues play.
Catalytic Residues Roles
| UniProt | PDB* (1n1d) | ||
| Lys44, Lys46 | Lys44A, Lys46A | Help stabilise the negatively charged transition state. | attractive charge-charge interaction, hydrogen bond donor, electrostatic stabiliser |
Chemical Components
bimolecular nucleophilic substitution, overall reactant used, overall product formed, native state of enzyme regenerated, rate-determining stepReferences
- Pattridge KA et al. (2003), J Biol Chem, 278, 51863-51871. Glycerol-3-phosphate Cytidylyltransferase: STRUCTURAL CHANGES INDUCED BY BINDING OF CDP-GLYCEROL AND THE ROLE OF LYSINE RESIDUES IN CATALYSIS. DOI:10.1074/jbc.m306174200. PMID:14506262.
- Mericl AN et al. (2012), Med Sci Monit, 18, BR427-BR434. Comparative kinetic analysis of glycerol 3-phosphate cytidylyltransferase from Enterococcus faecalis and Listeria monocytogenes. DOI:10.12659/MSM.883535.
- Weber CH et al. (1999), Structure, 7, 1113-1124. A prototypical cytidylyltransferase: CTP:glycerol-3-phosphate cytidylyltransferase from Bacillus subtilis. DOI:10.1016/s0969-2126(99)80178-6. PMID:10508782.
- Carter CW Jr (1993), Annu Rev Biochem, 62, 715-748. Cognition, Mechanism, and Evolutionary Relationships in Aminoacyl-tRNA Synthetases. DOI:10.1146/annurev.bi.62.070193.003435. PMID:8352600.

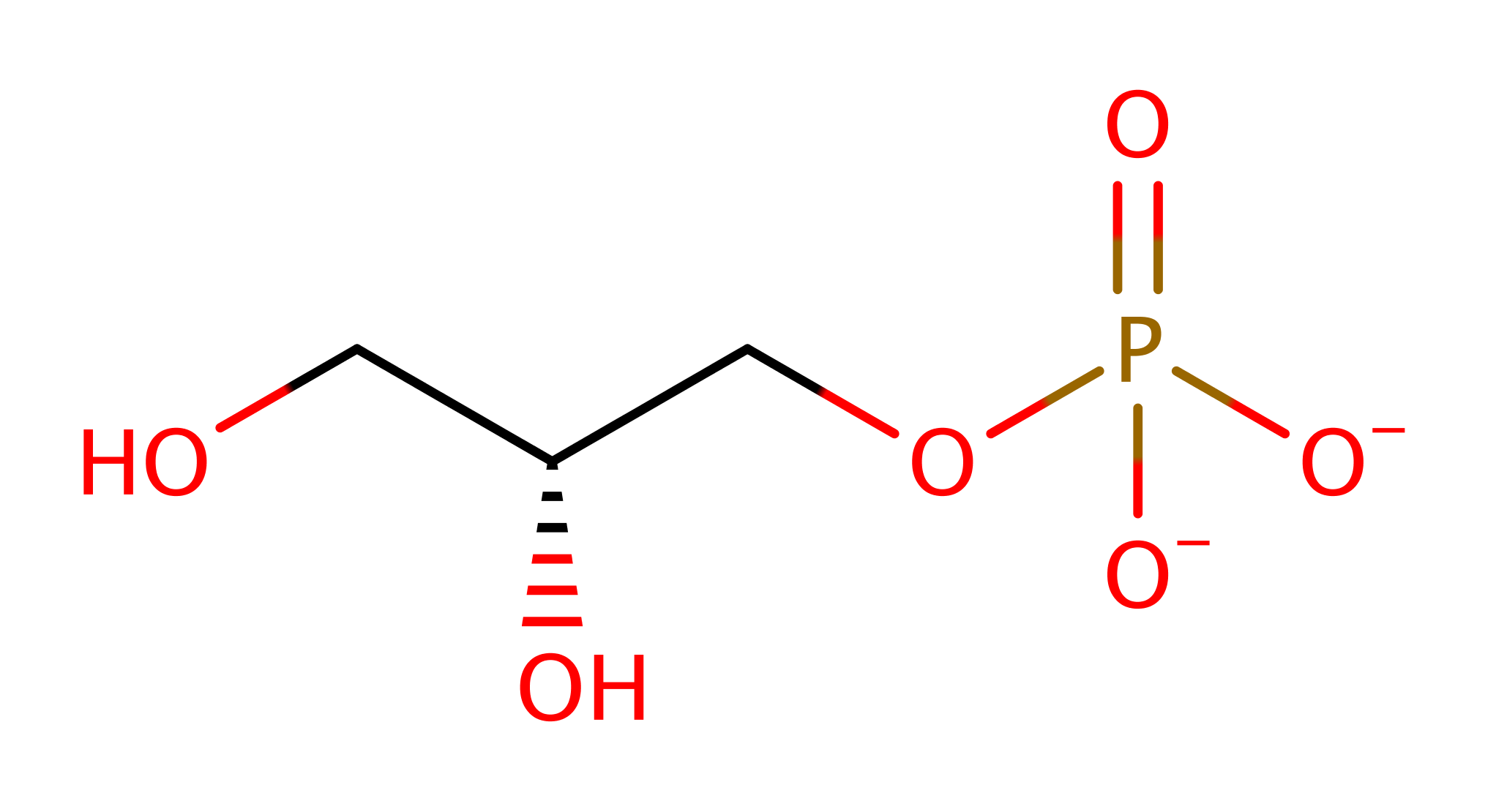
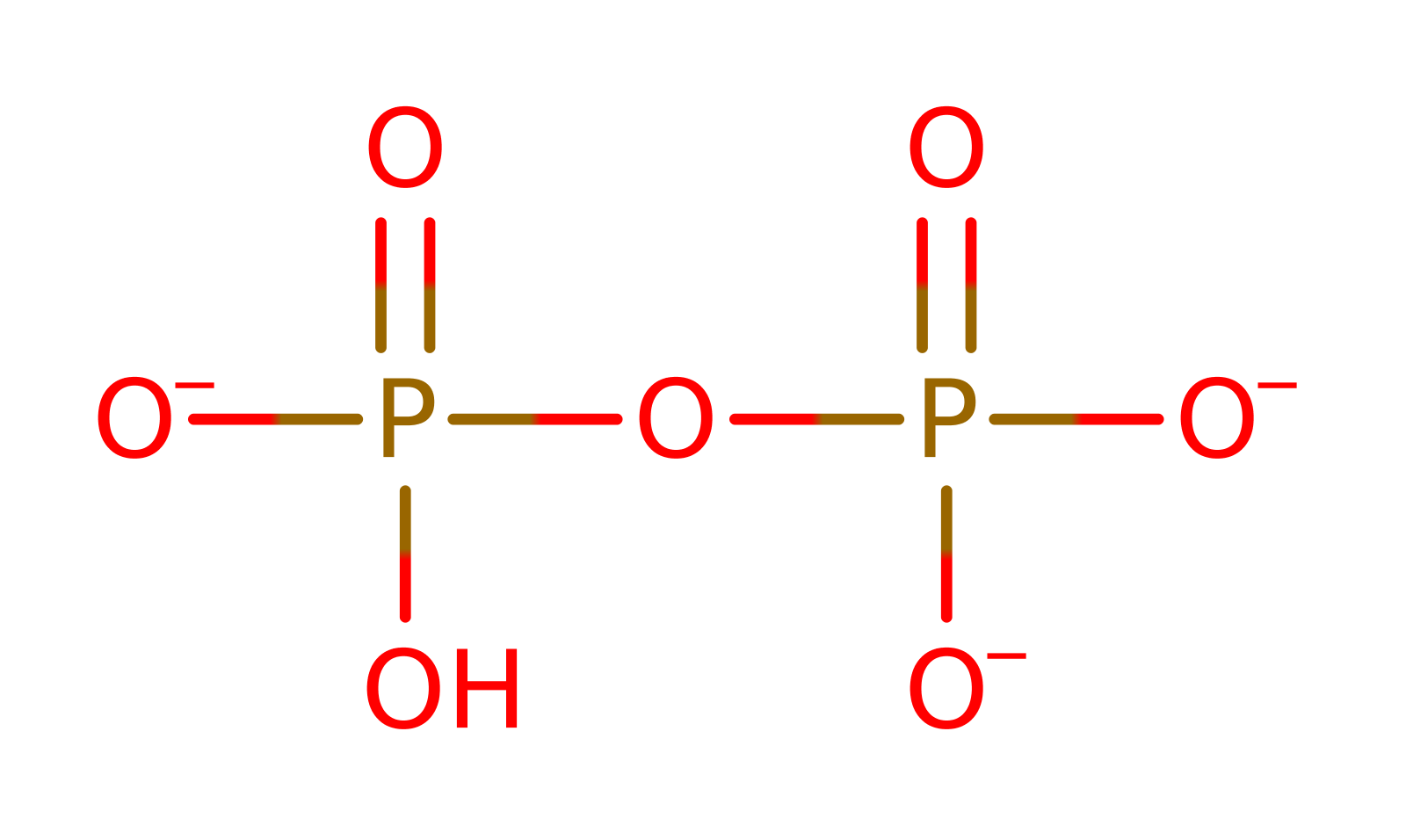
Step 1. The glycerol-3-phosphate initiates a nucleophilic attack on the alpha phosphate of CTP in a substitution reaction. . The magnesium enhances the electrophilicity of the CTP alpha phosphate by reducing the repulsion between this group and the attacking nucleophile. Kinetic studies have shown the divalent magnesium ion to be essential for catalysis. Pattridge et al. [PMID:14506262] suggests that the ion coordinates between the alpha phosphate of the CTP and the attacking phosphate of glycerol-3-phosphate [PMID:14506262].
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys46A | attractive charge-charge interaction, hydrogen bond donor, electrostatic stabiliser |
| Lys44A | attractive charge-charge interaction, hydrogen bond donor, electrostatic stabiliser |



 Download:
Download: