E1 ubiquitin-activating enzyme
Ubiquitination is mediated by three enzymes, E1 (PDB:3cmm), E2 (PDB:1ayz) and E3 (PDB:1c4z). The first, E1, is essential for ubiquitin activation and transferring the substrate onto the second cascade enzyme, E2, which responsible for mediating repeated ubiquitination at the eventual substrate, which is brought into proximity of E2 by the active site of E3, the enzyme which also regulates substrate specificity. This cascade regulates the ubiquitination of specific proteins, generating post-translational modifications that activate a number of possible cellular responses, depending upon the site and type of ubiquitin marker.
This entry represents the first of the three reactions occurring in this cascade: ubiquitin:[E1 ubiquitin-activating enzyme] ligase (AMP-forming). E1 catalyses the ATP-dependent activation of ubiquitin through the formation of a thioester bond between the C-terminal glycine of ubiquitin and the sulfhydryl side group of a cysteine residue in the E1 protein. Several E1 type enzymes have been identified to catalyse substrate-specific transfer reactions for ubiquitin-like proteins, although a number of these enzymes will also catalyse the transfer of ubiquitin and other ubiquitin-like proteins.
Reference Protein and Structure
- Sequences
-
P22515
 (6.2.1.45)
(6.2.1.45)
P06104 (2.3.2.23)
(2.3.2.23)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Saccharomyces cerevisiae S288c (Baker's yeast)

- PDB
-
3cmm
- Crystal Structure of the Uba1-Ubiquitin Complex
(2.7 Å)



- Catalytic CATH Domains
-
3.50.50.80
 1.10.10.2660
1.10.10.2660  3.40.50.720
3.40.50.720  (see all for 3cmm)
(see all for 3cmm)
- Cofactors
- Magnesium(2+) (1) Metal MACiE
Enzyme Reaction (EC:6.2.1.45)
Enzyme Mechanism
Introduction
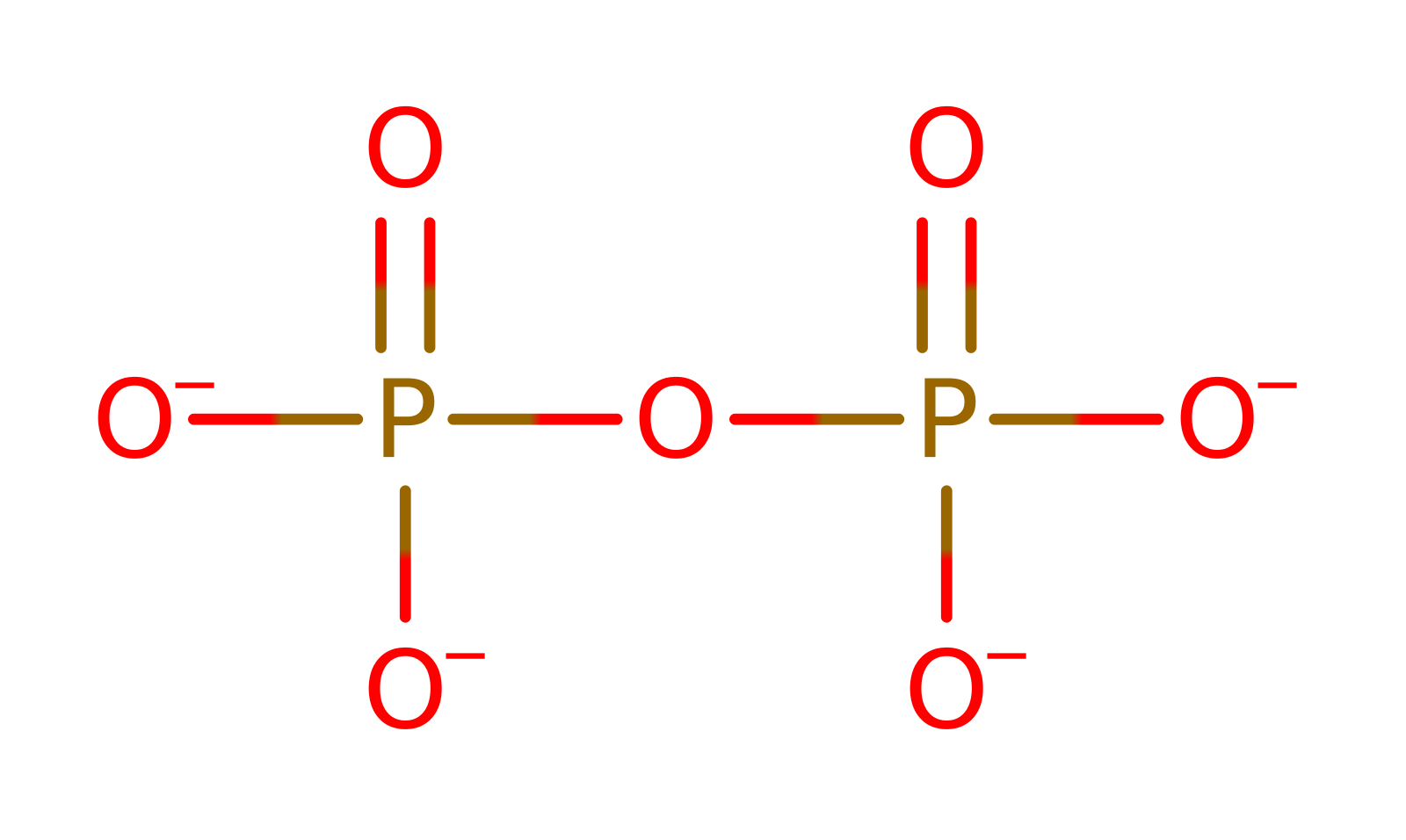
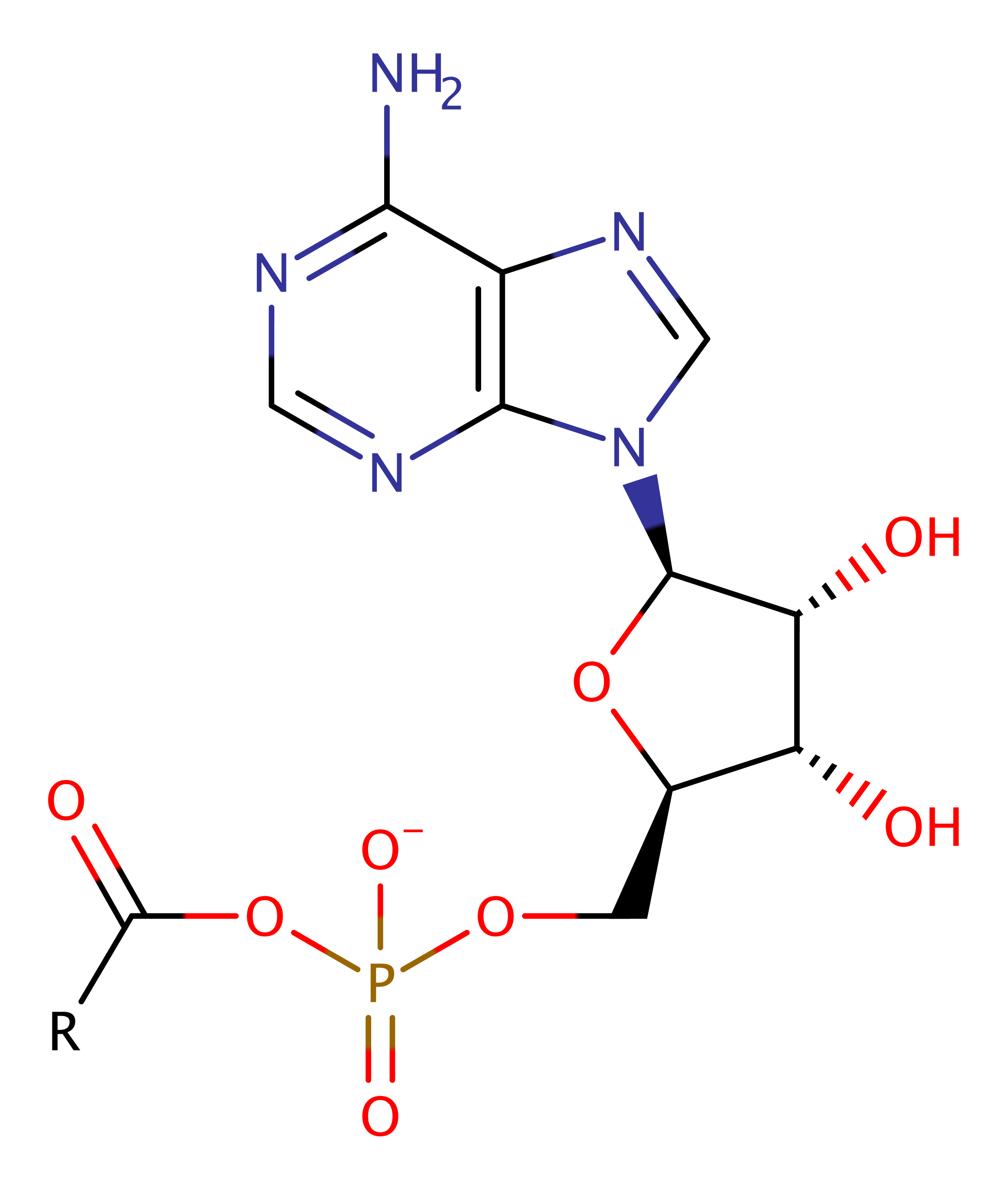
The two-step reaction consists of the ATP-dependent formation of an E1-ubiquitin adenylate intermediate in which the C-terminal glycine of ubiquitin is bound to AMP via an acyl-phosphate linkage, then followed by the conversion to an E1-ubiquitin thioester bond via the cysteine residue on E1 in the second step.
Catalytic Residues Roles
| UniProt | PDB* (3cmm) | ||
| Asn781 (main-N), Asp782 (main-N) | Asn781(772)A (main-N), Asp782(773)A (main-N) | Forms the oxyanion hole that stabilises the negatively charged intermediates and transition states involved in the transfer of the ubiquitin from AMP to Cys600A in E1. | hydrogen bond donor, electrostatic stabiliser |
| Thr601 | Thr601(592)A | Structural conservation and crystallographic data suggests that Thr601A may act as a general base, both towards Cys600A of E1 and also Cys88A of E2. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, proton relay, increase acidity, increase nucleophilicity |
| Asp544 | Asp544(535)A | Helps ensure the substrates are held in the correct positions for the reaction to occur, also destabilises the ground state during the ubiquitin adenylation steps. | steric role |
| Cys600 | Cys600(591)A | This is the E1 residue that becomes ubiqutinated. Also acts as a general acid/base in this portion of the reaction. | covalently attached, hydrogen bond donor, nucleophile, proton donor, activator |
| Arg481, Arg603, Arg21 | Arg481(472)A, Arg603(594)A, Arg21(12)A | Helps stabilise the reactive intermediates formed during the course of the reaction. | hydrogen bond donor, steric role, electrostatic stabiliser |
Chemical Components
bimolecular nucleophilic substitution, overall reactant used, overall product formed, intermediate formation, bimolecular nucleophilic addition, enzyme-substrate complex formation, unimolecular elimination by the conjugate base, intermediate collapseReferences
- Schulman BA et al. (2009), Nat Rev Mol Cell Biol, 10, 319-331. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. DOI:10.1038/nrm2673. PMID:19352404.
- Olsen SK et al. (2010), Nature, 463, 906-912. Active site remodelling accompanies thioester bond formation in the SUMO E1. DOI:10.1038/nature08765. PMID:20164921.
- Lee I et al. (2008), Cell, 134, 268-278. Structural Insights into E1-Catalyzed Ubiquitin Activation and Transfer to Conjugating Enzymes. DOI:10.1016/j.cell.2008.05.046. PMID:18662542.
- Capili AD et al. (2007), Curr Opin Struct Biol, 17, 726-735. Taking it step by step: mechanistic insights from structural studies of ubiquitin/ubiquitin-like protein modification pathways. DOI:10.1016/j.sbi.2007.08.018. PMID:17919899.
- Passmore LA et al. (2004), Biochem J, 379, 513-525. Getting into position: the catalytic mechanisms of protein ubiquitylation. DOI:10.1042/bj20040198. PMID:14998368.
- Pickart CM et al. (2004), Biochim Biophys Acta, 1695, 55-72. Ubiquitin: structures, functions, mechanisms. DOI:10.1016/j.bbamcr.2004.09.019. PMID:15571809.
- Wu PY et al. (2003), EMBO J, 22, 5241-5250. A conserved catalytic residue in the ubiquitin-conjugating enzyme family. DOI:10.1093/emboj/cdg501. PMID:14517261.
- Lake MW et al. (2001), Nature, 414, 325-329. Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex. DOI:10.1038/35104586. PMID:11713534.
- Scheffner M et al. (1995), Nature, 373, 81-83. Protein ubiquitination involving an E1–E2–E3 enzyme ubiquitin thioester cascade. DOI:10.1038/373081a0. PMID:7800044.

Step 1. The catalytic residues in the adenylation site are thought to induce a reactive conformation in ATP, promoting nucleophilic attack from the ubiquitin C terminus, as well as stabilising the pentavalent transition state [PMID:19352404, PMID:15571809]. The C terminus of ubiquitin attacks the alpha phosphate of ATP within the adenylation binding site.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg481(472)A | steric role, hydrogen bond donor, electrostatic stabiliser |
| Arg21(12)A | steric role, hydrogen bond donor, electrostatic stabiliser |
| Asp544(535)A | steric role |
Chemical Components
ingold: bimolecular nucleophilic substitution, overall reactant used, overall product formed, intermediate formation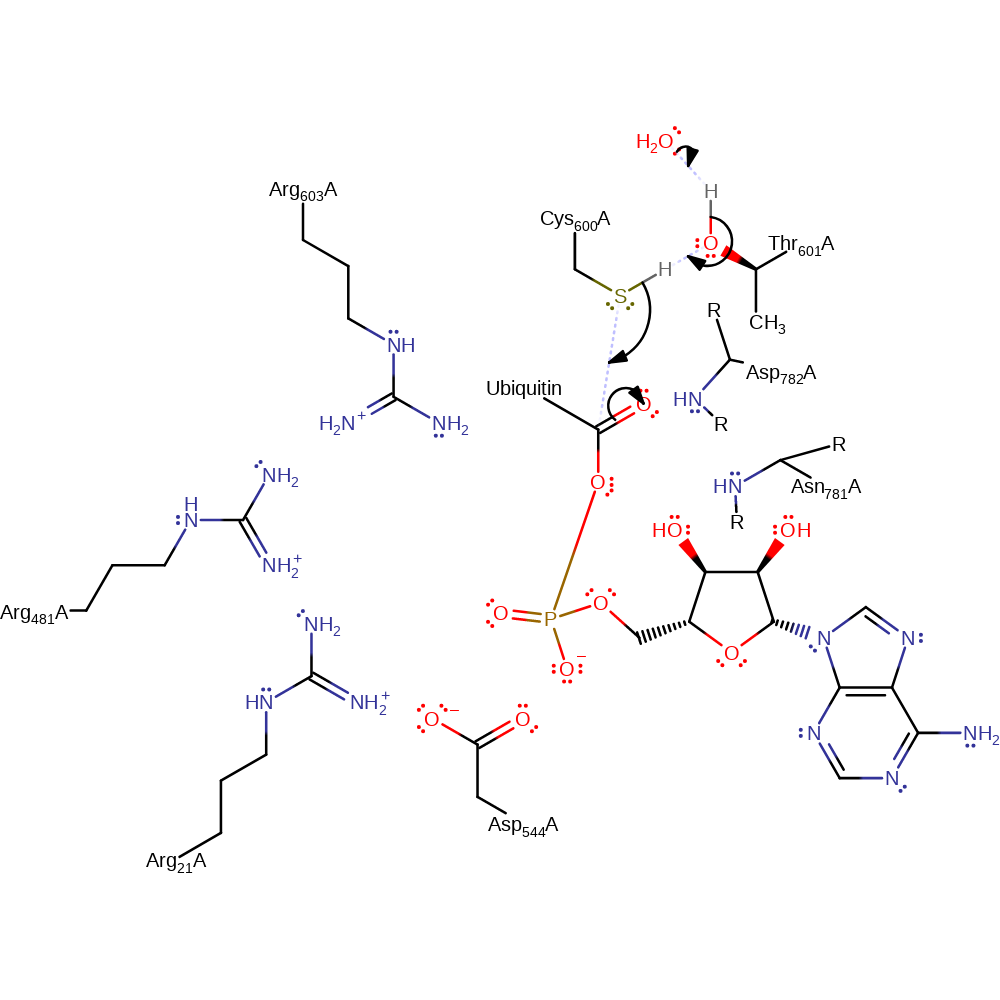
Step 2. The adenylation site is approx. 35 A from the catalytic cysteine, situated in the thio-ester formation site. A change in conformation upon adenylation is thought to bring the two reaction sites closer. This structural rearrangement is also thought to orientate residues into a catalytically active arrangement, including the postulated general base [PMID:19352404] that activates Cys600A towards nucleophilic attack at the adenylated ubiquitin.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg603(594)A | hydrogen bond donor, electrostatic stabiliser |
| Thr601(592)A | increase acidity, increase nucleophilicity, hydrogen bond acceptor |
| Asp782(773)A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Asn781(772)A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Cys600(591)A | hydrogen bond donor |
| Thr601(592)A | proton acceptor |
| Cys600(591)A | proton donor |
| Thr601(592)A | proton donor |
| Cys600(591)A | nucleophile |
| Thr601(592)A | proton relay |
Chemical Components
ingold: bimolecular nucleophilic addition, intermediate formation, enzyme-substrate complex formation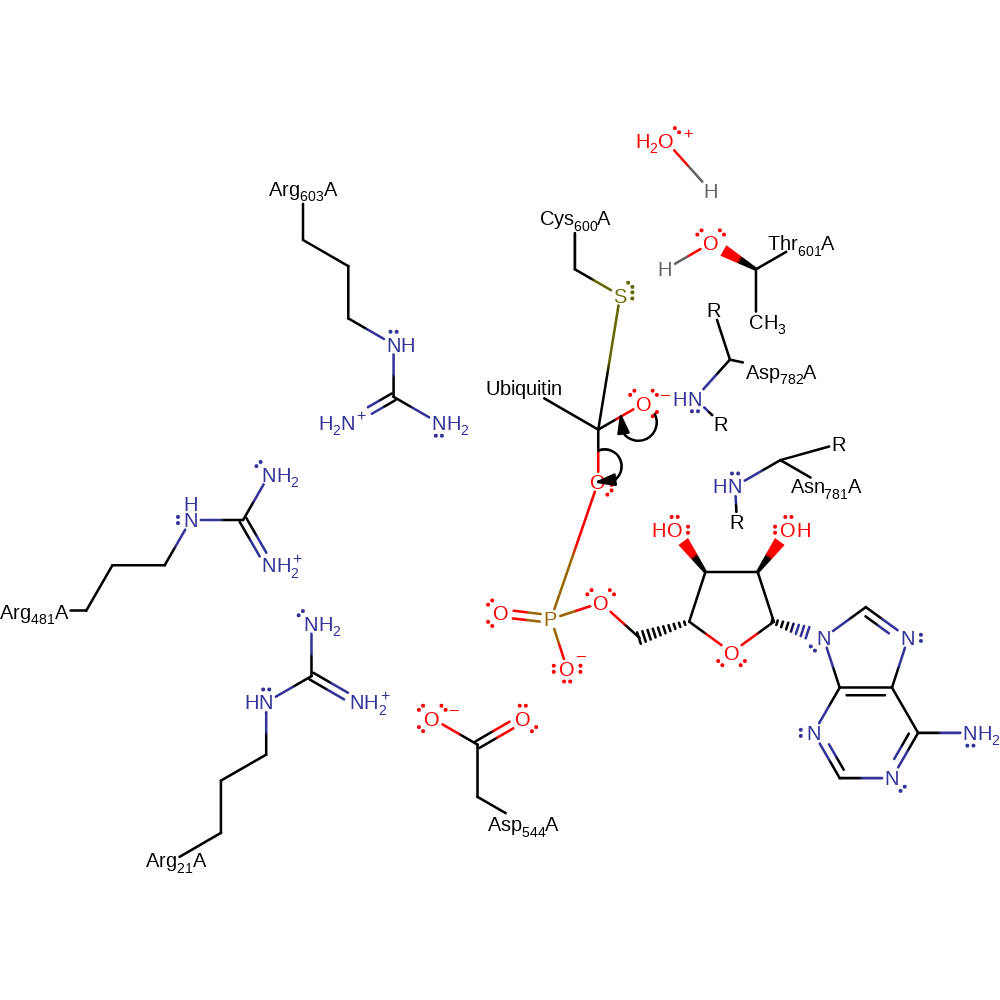
Step 3. The tetrahedral anionic intermediate collapses, releasing AMP and forming the enzyme-substrate adduct.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg603(594)A | hydrogen bond donor, electrostatic stabiliser |
| Thr601(592)A | hydrogen bond acceptor, hydrogen bond donor |
| Asp782(773)A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Asn781(772)A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Cys600(591)A | covalently attached, activator |
Chemical Components
ingold: unimolecular elimination by the conjugate base, intermediate collapse, intermediate formation
Step 4. A second round of ubiquitin adenylation is coupled to the formation of the high energy, activated enzyme bound ubiquitin intermediates. The C terminus of a second ubiquitin attacks the alpha phosphate of ATP within the adenylation binding site.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg481(472)A | steric role, hydrogen bond donor, electrostatic stabiliser |
| Arg21(12)A | steric role, hydrogen bond donor, electrostatic stabiliser |
| Asp544(535)A | steric role |
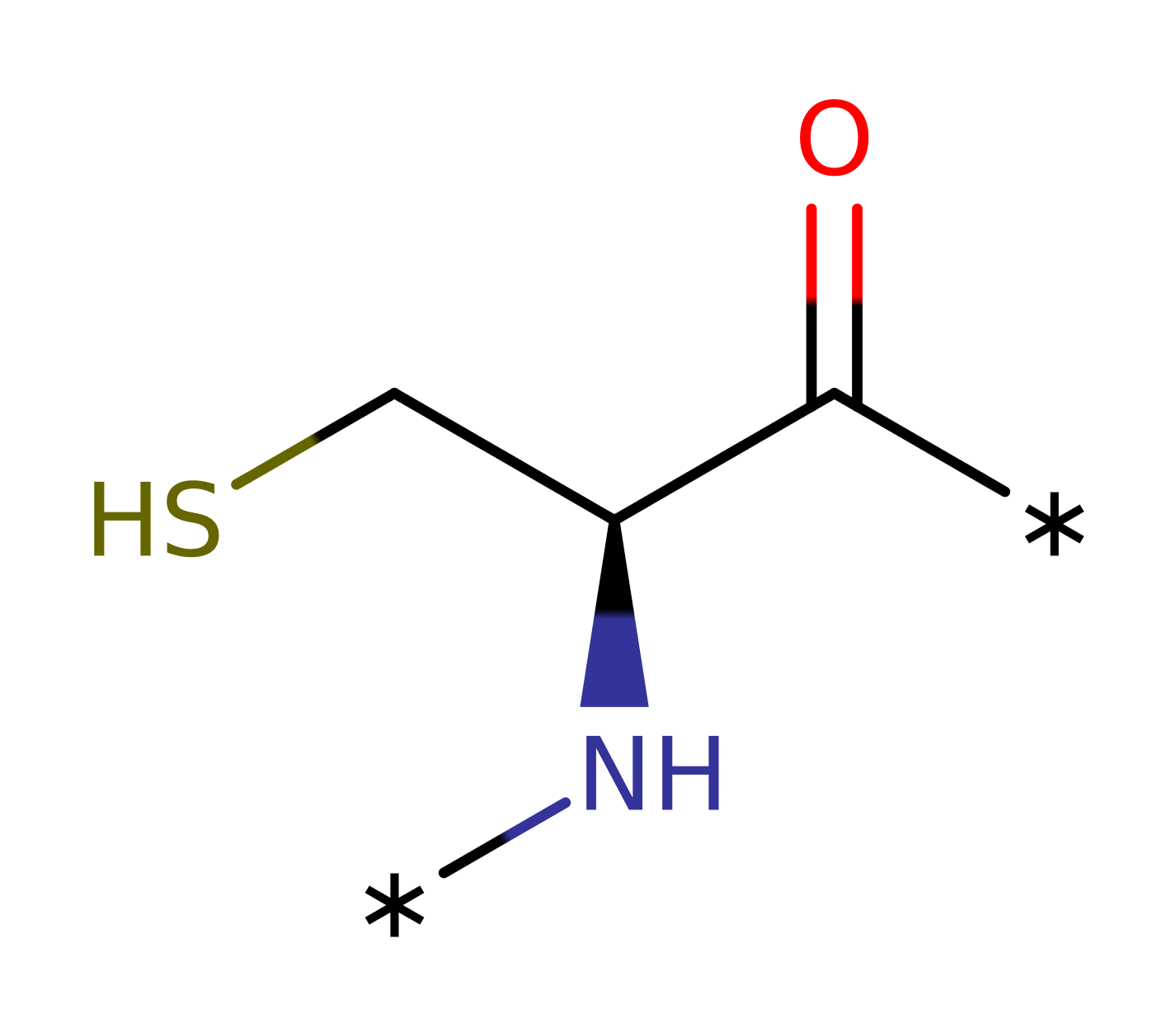



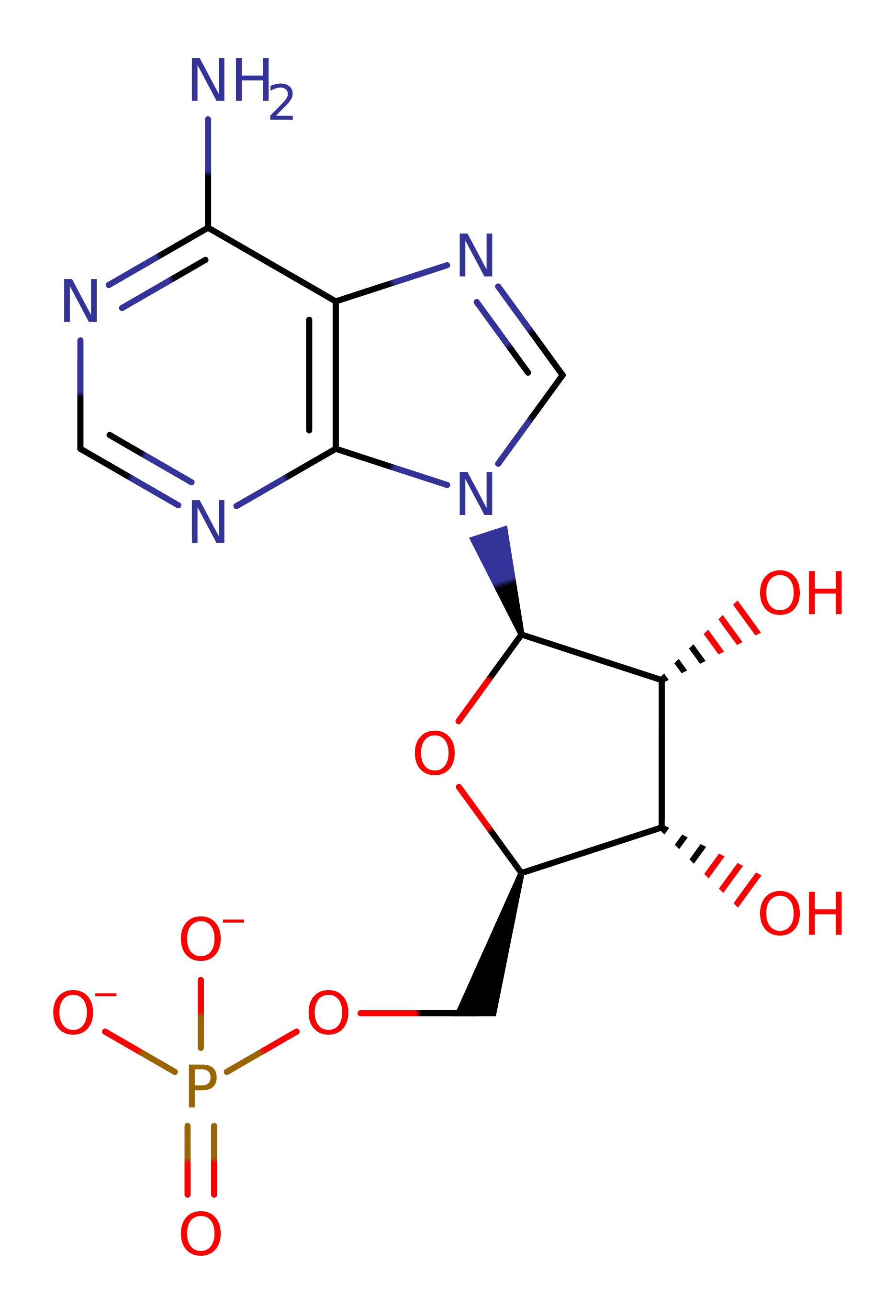
 Download:
Download: