Isoleucine-tRNA ligase (type 2)
Isoleucine-tRNA ligase (also known as Isoleucyl-tRNA synthetase) is an alpha monomer that belongs to class Ia tRNA synthetases. It activates not only the cognate substrate L-isoleucine but also the structurally similar L-valine in the first, amino-acylation step. In a second, "editing" step, the synthetase itself rapidly hydrolyses only the valylated products. For this two-step substrate selection, a "double-sieve" mechanism has been proposed to explain the high substrate fidelity and low copy error of the tRNA synthetase.
Escherichia coli isoleucyl-tRNA synthetasehas been shown to contain two enzyme-bound zinc atoms per polypeptide chain. The N-terminal zinc ion may play a structural role. Although the two zinc domains are always conserved, the zinc ligands are not, so that there can be both zinc atoms, no zinc atoms or only one of the two. The C-terminal zinc ion is essential (for function in vivo). It plays an important roles in aminoacylation of tRNA(Ile) [PMID:7488160, PMID:8672449].
Reference Protein and Structure
- Sequence
-
P56690
 (6.1.1.5)
(6.1.1.5)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Thermus thermophilus HB8 (Bacteria)

- PDB
-
1ile
- ISOLEUCYL-TRNA SYNTHETASE
(2.5 Å)



- Catalytic CATH Domains
-
3.40.50.620
 (see all for 1ile)
(see all for 1ile)
- Cofactors
- Magnesium(2+) (1), Zinc(2+) (1) Metal MACiE
Enzyme Reaction (EC:6.1.1.5)
Enzyme Mechanism
Introduction
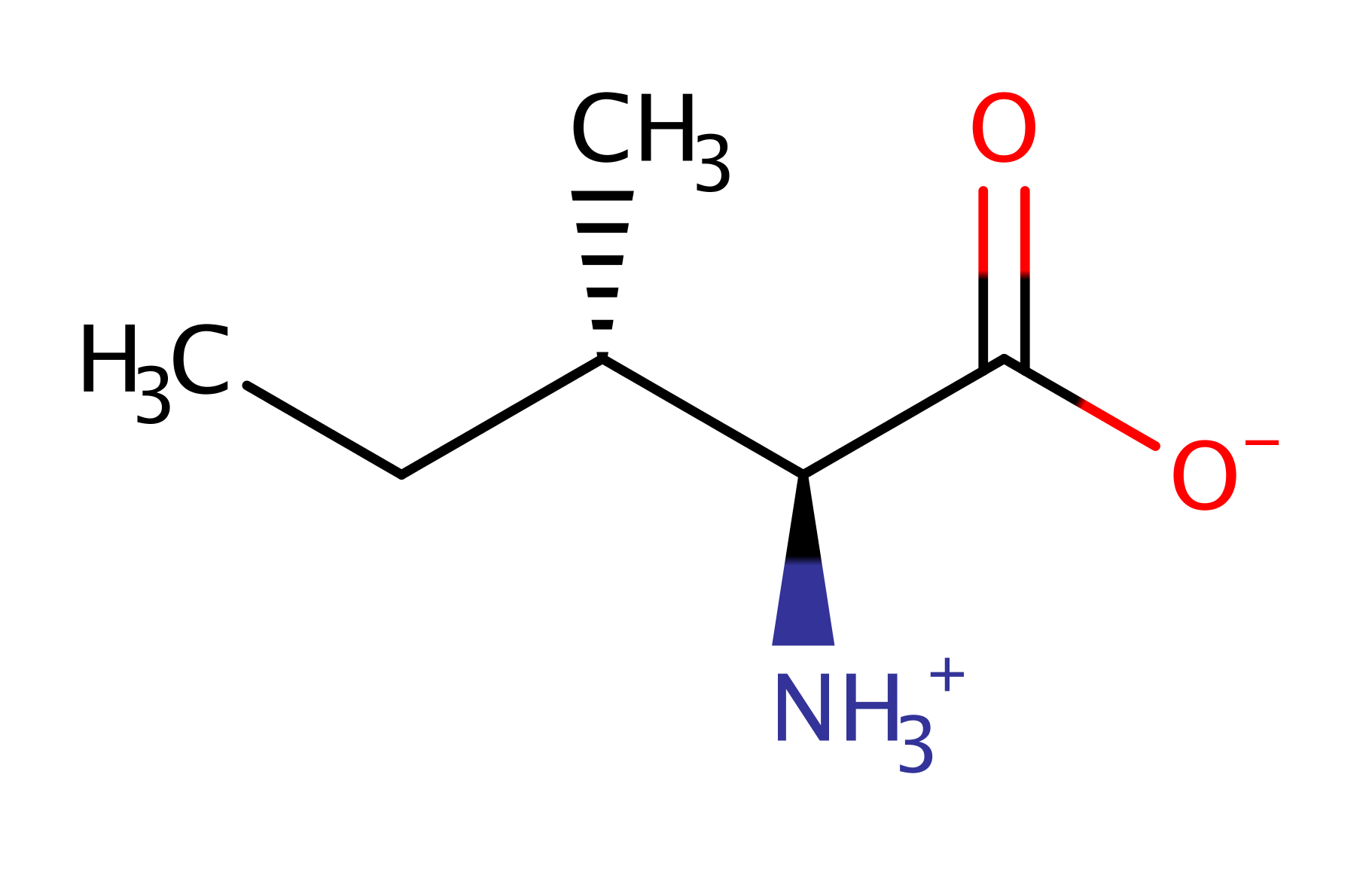
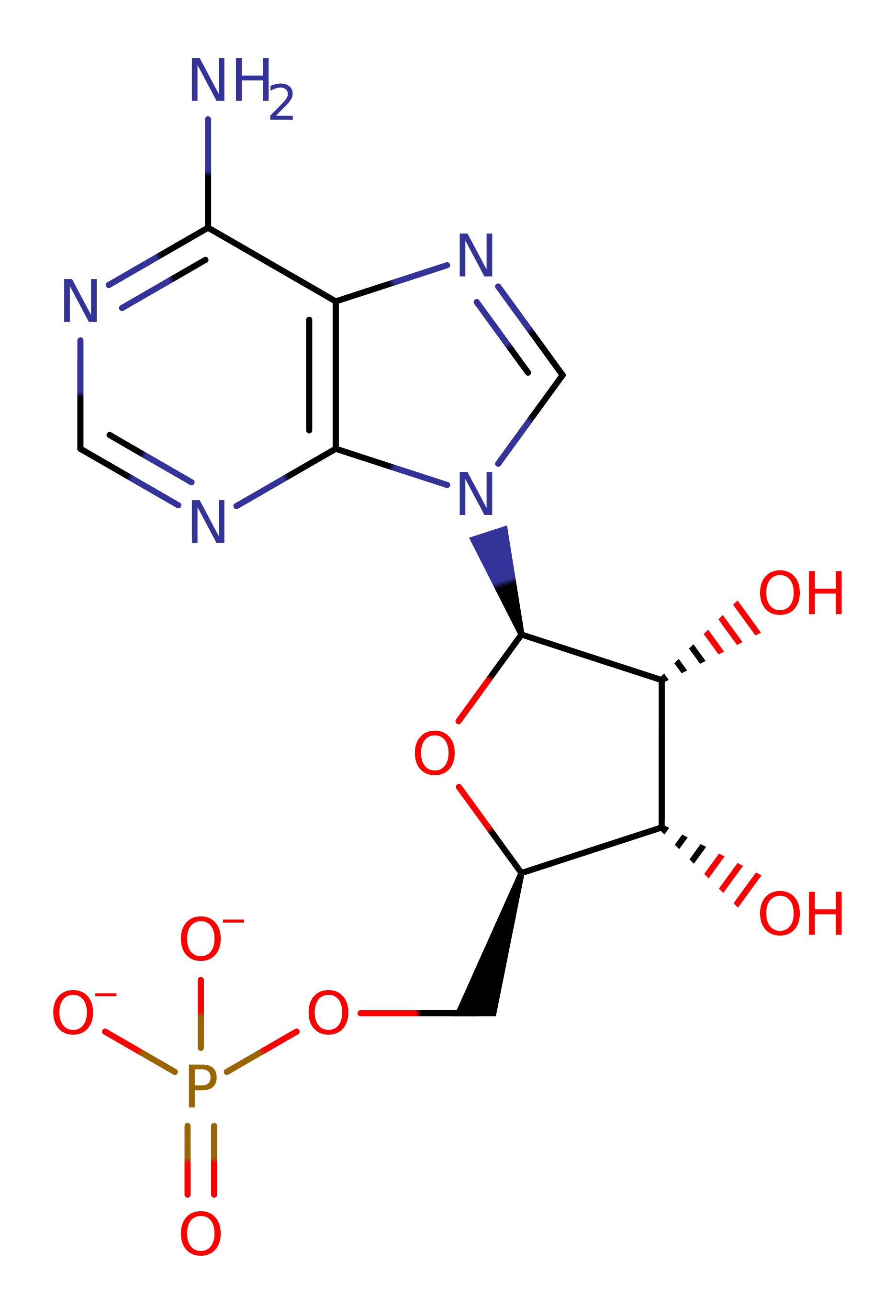
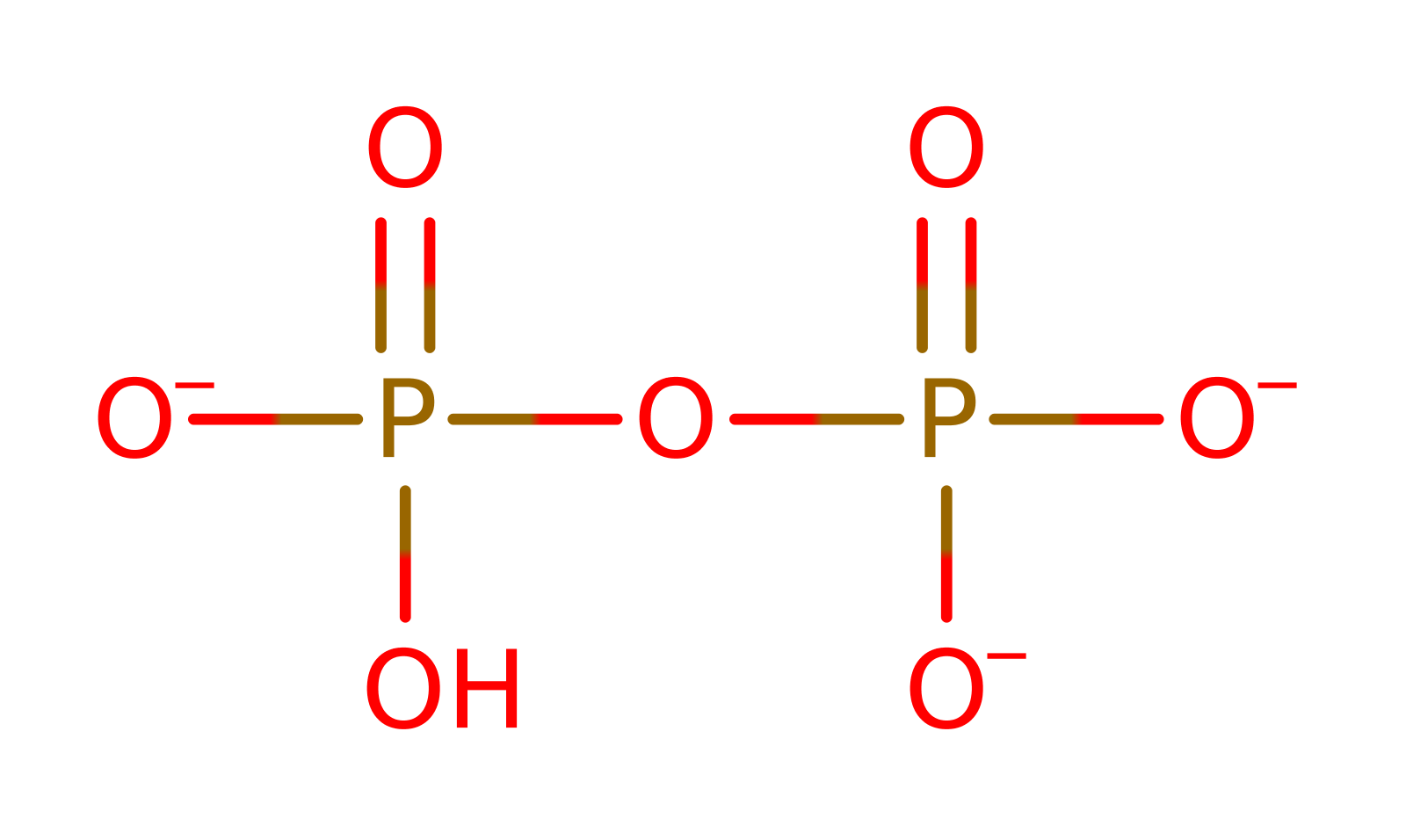
The zwitterionic leucine substrate attacks the alpha phosphate of ATP, forming an activated, adenylated, amino acid. The 2' end of the specific tRNA molecule attacks at the activated phospho-ester bond. The anionic, tetrahedral intermediate collapses, generating the tRNA-isoleucine adduct and AMP.
Catalytic Residues Roles
| UniProt | PDB* (1ile) | ||
| Trp518, Trp558 | Trp518A, Trp558A | The bulky tryptophan residues prevent the binding of larger amino acids within the adenylation site. These residues form the first molecular sieve, introducing rough substrate specificity between large and small amino acids [PMID:9554847, PMID:10966471] | van der waals interaction, steric role |
| Lys591, Lys594 | Lys591A, Lys594A | Help stabilise the reactive intermediates formed during the course of the reaction. | attractive charge-charge interaction, hydrogen bond donor, electrostatic stabiliser |
| Gln554, Asp85, Pro46 | Gln554A, Asp85A, Pro46A | Form the second molecular sieve site, which discriminates between wrongly activated amino acids, such as valine and the correct substrate for father catalysis. | hydrogen bond acceptor, steric role |
Chemical Components
bimolecular nucleophilic substitution, overall reactant used, overall product formed, intermediate formation, bimolecular nucleophilic addition, proton transfer, rate-determining step, unimolecular elimination by the conjugate base, intermediate collapse, native state of enzyme regeneratedReferences
- Nureki O et al. (1998), Science, 280, 578-582. Enzyme Structure with Two Catalytic Sites for Double-Sieve Selection of Substrate. DOI:10.1126/science.280.5363.578. PMID:9554847.
- Fukai S et al. (2003), RNA, 9, 100-111. Mechanism of molecular interactions for tRNAVal recognition by valyl-tRNA synthetase. DOI:10.1261/rna.2760703. PMID:12554880.
- Ibba M et al. (2000), Annu Rev Biochem, 69, 617-650. Aminoacyl-tRNA Synthesis. DOI:10.1146/annurev.biochem.69.1.617. PMID:10966471.
- Fersht AR (1977), Biochemistry, 16, 1025-1030. Editing mechanisms in protein synthesis. Rejection of valine by the isoleucyl-tRNA synthetase. DOI:10.1021/bi00624a034. PMID:321008.

Step 1. The bulky tryptophan residues prevent the binding of larger amino acids within the adenylation site. The zwitterionic leucine substrate attacks the alpha phosphate of ATP, forming an activated, adenylated, amino acid.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp85A | attractive charge-charge interaction, steric role, hydrogen bond acceptor |
| Trp518A | steric role, van der waals interaction |
| Lys591A | attractive charge-charge interaction, electrostatic stabiliser, hydrogen bond donor |
| Pro46A | steric role, van der waals interaction |
| Lys594A | attractive charge-charge interaction, electrostatic stabiliser, hydrogen bond donor |
| Trp558A | steric role, van der waals interaction |
| Gln554A | steric role, hydrogen bond acceptor |
Chemical Components
ingold: bimolecular nucleophilic substitution, overall reactant used, overall product formed, intermediate formation
Step 2. Amino acids similar in size to isoleucine which slip through the first 'sieve' and are mistakenly adenylated, such as valine, are channelled to the site of the second, finer molecular sieve. This site uses a precise molecular architecture to discriminate between wrongly activated amino acids, such as valine and the correct substrate for father catalysis. Once bound within this second site, catalytically activated hydrolysis deactivates the amino acid, preventing it from attaching to the isoleucine tRNA. The residue Thr232 is thought to be responsible for amino acid recognition at this site. This two sieve mechanism further reduces the risk of missense mutations in later protein synthesis [PMID:9554847, PMID:12554880, PMID:321008]. In this step, the 2' end of the specific tRNA molecule attacks at the activated phospho-ester bond
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp85A | attractive charge-charge interaction, electrostatic stabiliser, hydrogen bond acceptor |
| Trp518A | van der waals interaction |
| Lys591A | attractive charge-charge interaction, electrostatic stabiliser, hydrogen bond donor |
| Pro46A | van der waals interaction |
| Lys594A | attractive charge-charge interaction, electrostatic stabiliser, hydrogen bond donor |
| Trp558A | van der waals interaction |
| Gln554A | hydrogen bond acceptor |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer, intermediate formation, overall reactant used, rate-determining step
Step 3. The anionic, tetrahedral intermediate collapses, generating the tRNA-isoleucine adduct and AMP.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp85A | hydrogen bond acceptor |
| Trp518A | van der waals interaction |
| Lys591A | attractive charge-charge interaction, electrostatic stabiliser, hydrogen bond donor |
| Pro46A | van der waals interaction |
| Lys594A | attractive charge-charge interaction, electrostatic stabiliser, hydrogen bond donor |
| Trp558A | van der waals interaction |
| Gln554A | hydrogen bond acceptor |



 Download:
Download: