DNA-3-methyladenine glycosylase II
A variety of environmental toxins can react with DNA and chemically alkylate the bases. This blocks the replicative polymerases and interferes with binding of regulatory proteins to DNA resulting in widespread cellular responses including activation of cell cycle checkpoints and programmed cell death. There are a number of repair strategies - direct reversal, nucleotide excision repair and base excision repair. Most single base modifications are corrected using base excision repair. Firstly a lesion specific DNA glycosylase removes the damaged base by hydrolysis of the deoxyribose glycosidic bond of alkylated DNA. The abasic site is then excised and filled by DNA polymerase.
AlkA lacks an obvious residue that could fulfill the role of a general acid to protonate the leaving group and later activate a water molecule. It is thought that AlkA's positively charged, alkylated substrates might not require this type of assistance. The lack of a general acid in AlkA's active site provides a means of catalytic selectivity. Positively charged bases, which do not require protonation and have a weakened glycosylic bond, can be removed effectively, and in turn it may function as a general base to activate a nearby water molecule. Unmodified bases require protonation by a general acid prior to hydrolysis and therefore are poor substrates. [PMID:10675345]
Reference Protein and Structure
- Sequence
-
P04395
 (3.2.2.21)
(3.2.2.21)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1diz
- CRYSTAL STRUCTURE OF E. COLI 3-METHYLADENINE DNA GLYCOSYLASE (ALKA) COMPLEXED WITH DNA
(2.5 Å)



- Catalytic CATH Domains
-
1.10.1670.10
 1.10.340.30
1.10.340.30  (see all for 1diz)
(see all for 1diz)
Enzyme Reaction (EC:3.2.2.21)
Enzyme Mechanism
Introduction
The bound DNA is highly distorted. This is achieved by surprisingly few contacts. Lys170 interacts with the DNA phosphate backbone. The active site cleft is flanked by a wedge tipped with Leu125 which juts into the minor groove and displaces the base targeted for excision. It also causes a 66 degree bend in the DNA away from the protein which disrupts base pair stacking interactions either side of the flipped out base. At the base of the wedge Pro175 is forced into the minor groove and makes Van der Waals contacts with the deoxyribose sugar leading to a two fold expansion of the minor groove. The DNA is tightly anchored to the protein by a series of hydrogen bonds and metal mediated interaction by residues 202 to 227. Gly214, Gly216 and Thr219 are involved in hydrogen bonding whilst the main chain carbonyl atoms of Gln210, Phe212 and Ile215 along with the phosphate oxygen of the DNA backbone and a water molecule coordinate a metal ion, thought to be sodium. Although the reaction could proceed via an Sn2 mechanism the lack of space present for a water nucleophile suggests an Sn1 type reaction. The positive nature of the substrate means that they do not require a general acid to protonate them. Since there is no suitably placed general acid this explains the selectivity for positive modified bases.
Catalytic Residues Roles
| UniProt | PDB* (1diz) | ||
| Asp238 | Asp238A(E) | The carboxylate group of Asp238 stabilises the carbocation intermediate by ionic interactions leading to the release of the alkylated base. | attractive charge-charge interaction, promote heterolysis, electrostatic stabiliser |
| Tyr222, Trp218 | Tyr222A(E), Trp218A(E) | The active site cleft is lined with electron rich aromatic rings from Phe18, Trp218, Tyr222, Trp272 and Tyr273 which form strong pi-donor/acceptor interactions with the electron deficient substrate, stabilising the extrahelical conformation of the alkylated base. | van der waals interaction, electrostatic stabiliser |
Chemical Components
heterolysis, overall reactant used, intermediate formation, proton transfer, bimolecular nucleophilic addition, intermediate terminated, overall product formedReferences
- Hollis T et al. (2000), Mutat Res, 460, 201-210. Structural studies of human alkyladenine glycosylase and E. coli 3-methyladenine glycosylase. DOI:10.1016/s0921-8777(00)00027-6. PMID:10946229.
- Hollis T et al. (2000), EMBO J, 19, 758-766. DNA bending and a flip-out mechanism for base excision by the helix–hairpin–helix DNA glycosylase, Escherichia coli AlkA. DOI:10.1093/emboj/19.4.758. PMID:10675345.
- Labahn J et al. (1996), Cell, 86, 321-329. Structural Basis for the Excision Repair of Alkylation-Damaged DNA. DOI:10.1016/s0092-8674(00)80103-8. PMID:8706136.

Step 1. The DNA substrate undergoes heterolysis, eliminating the alkylated base. This is the first step in the SN1 reaction, the heterolytic cleavage of the weakened glycosidic bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp238A(E) | attractive charge-charge interaction, promote heterolysis, electrostatic stabiliser |
| Tyr222A(E) | van der waals interaction, electrostatic stabiliser |
| Trp218A(E) | electrostatic stabiliser, van der waals interaction |
Chemical Components
heterolysis, overall reactant used, intermediate formationCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp238A(E) | attractive charge-charge interaction, electrostatic stabiliser |
| Tyr222A(E) | van der waals interaction, electrostatic stabiliser |
| Trp218A(E) | electrostatic stabiliser, van der waals interaction |


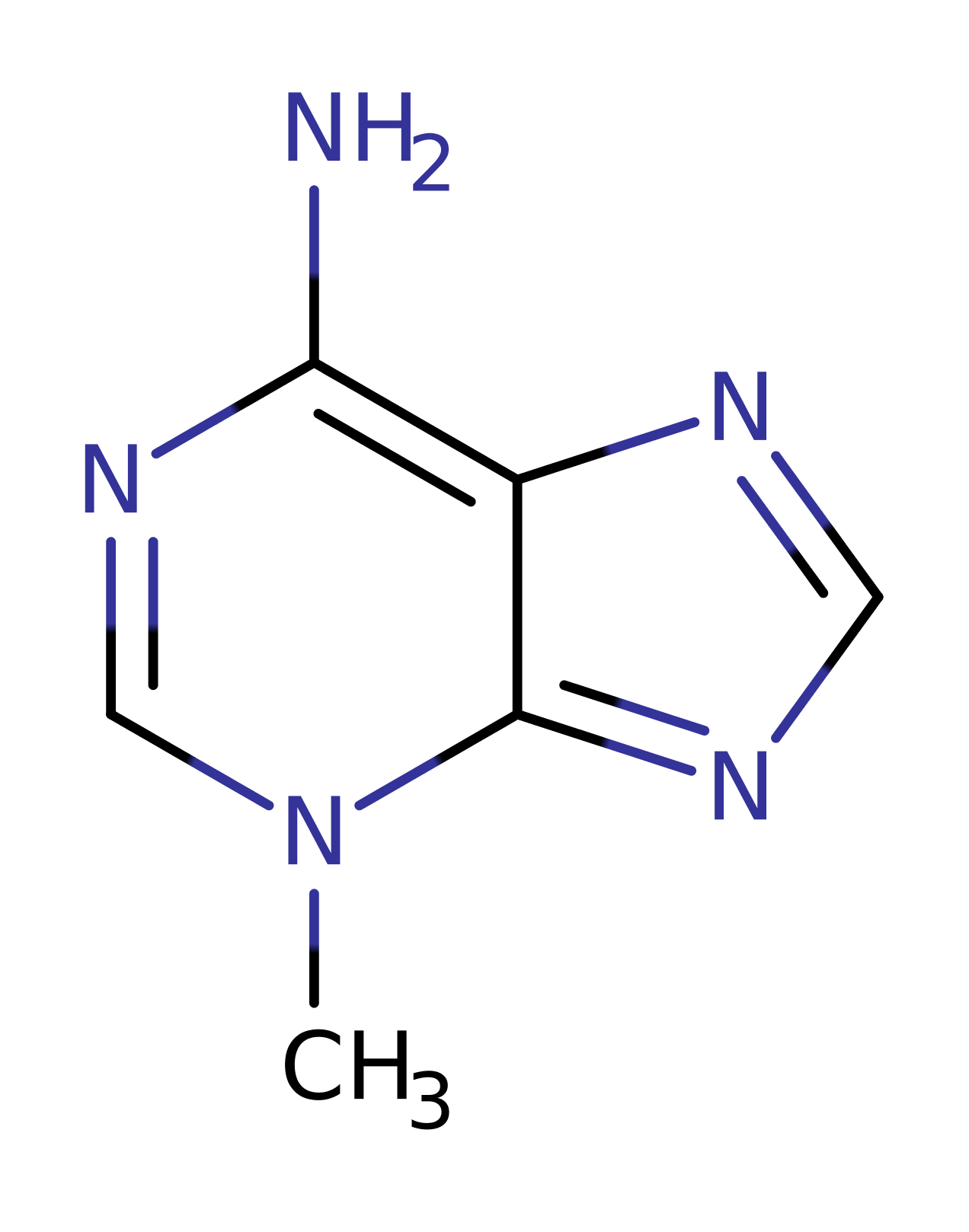



 Download:
Download: