Anthranilate synthase
Anthranilate synthase catalyses the initial reaction in tryptophan biosynthesis in microorganisms and plants. This enzyme is one of a family of glutamine amidotransferases, enzymes that utilise the amide of glutamine in the biosynthesis of amino acids, nucleotides, coenzymes, and an amino sugar. Glutamine amidotransferases thus exert major role in utilisation of assimilated nitrogen. Anthranilate synthase is the most thoroughly characterised glutamine amidotransferase.
The enzyme from Serratia marcescens is a heterotetramer of anthranilate synthase (TrpE) and glutamine amidotransferase (TrpG) subunits both of which are required for function.
Reference Protein and Structure
- Sequences
-
Q06128
 (4.1.3.27)
(4.1.3.27)
Q06129 (4.1.3.27)
(4.1.3.27)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Sulfolobus solfataricus P2 (Archaea)

- PDB
-
1qdl
- THE CRYSTAL STRUCTURE OF ANTHRANILATE SYNTHASE FROM SULFOLOBUS SOLFATARICUS
(2.5 Å)



- Catalytic CATH Domains
-
3.60.120.10
 3.40.50.880
3.40.50.880  (see all for 1qdl)
(see all for 1qdl)
- Cofactors
- Magnesium(2+) (1) Metal MACiE
Enzyme Reaction (EC:4.1.3.27)
Enzyme Mechanism
Introduction
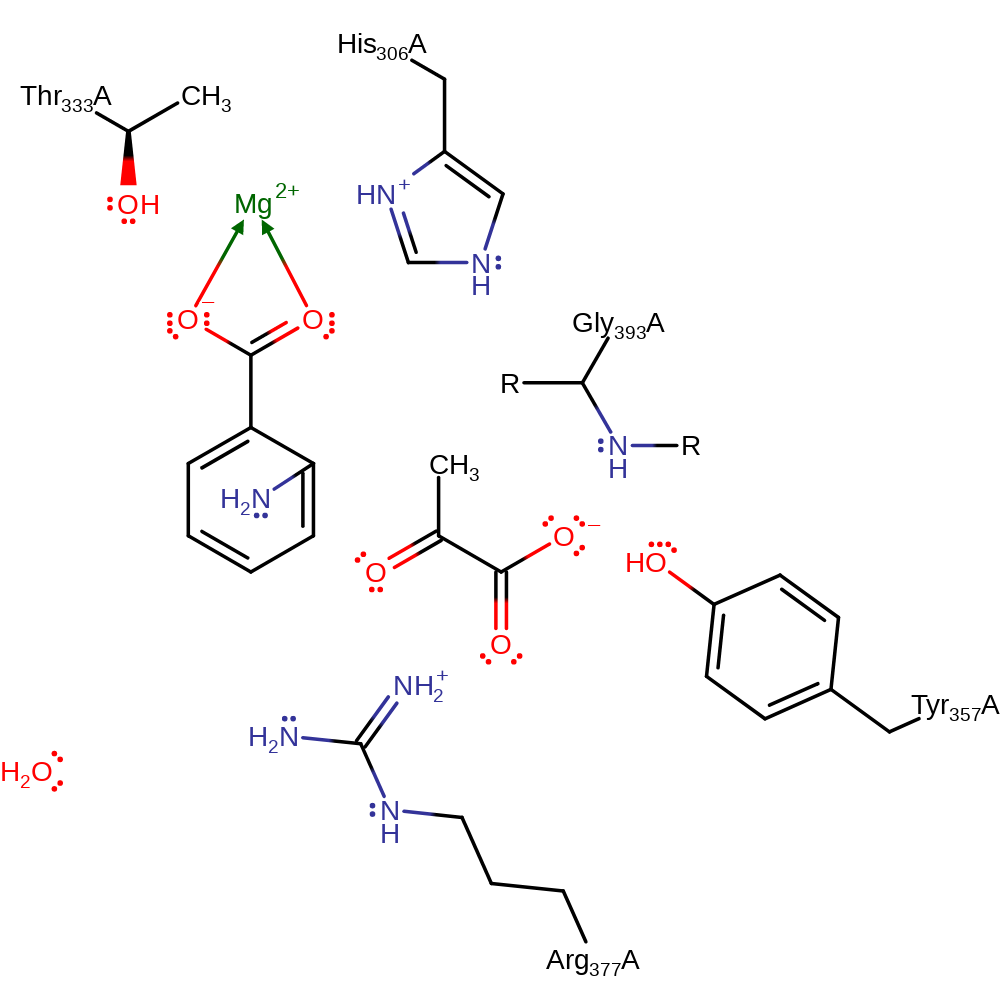
In the TrpG subunit anthranilate synthase produces anthranilate from chorismate via transfer of an amino group, (generated from glutamine) using a catalytic triad of with well-known mechanism consisting of Cys84, His175, and Glu177. In the second AIDC lyase part of the reaction a standard second-order elimination can be invoked to yield the double bond between C2 and C3 with a histidine (H306) as the base abstracting the C2 proton and pyruvate as the leaving group. Mg(II) and water provide an assisting acid group.
Catalytic Residues Roles
| UniProt | PDB* (1qdl) | ||
| Leu85 (main-N), Gly56 (main-N) | Leu85B (main-N), Gly56B (main-N) | Forms the oxyanion hole in the glutaminase domain. | hydrogen bond donor, electrostatic stabiliser |
| Cys84 | Cys84B | Acts to form a tetrahedral intermediate by attack on the substrate glutamine carbonyl group, subsequent breakdown of this releases ammonia which is transported to the second active site and amminates chorismate to form ADIC. | covalently attached, hydrogen bond acceptor, hydrogen bond donor, nucleophile, nucleofuge, proton donor, proton acceptor |
| His175 | His175B | Increases nucleophilicity of Cys85 via deprotonation of thiol side chain. Subsequently protonates the ammonia leaving group. In the active site regeneration hydrolysis, it deprotonates the substrate water, before reprotonating the thiol leaving group. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| His306, Thr333, Tyr357, Arg377, Gly393 (main-N) | His306(307)A, Thr333(334)A, Tyr357(358)A, Arg377(378)A, Gly393(394)A (main-N) | Act to stabilise the reactive intermediates and also to hold the chorismate substrate in the correct conformation. | proton relay, hydrogen bond acceptor, proton acceptor, proton donor |
| Glu177 | Glu177B | Acts to stabilise and activate the histidine (of the Cys-His-Glu triad) via hydrogen bonding. | increase basicity, hydrogen bond acceptor, increase acidity |
Chemical Components
proton transfer, bimolecular nucleophilic addition, enzyme-substrate complex formation, intermediate formation, overall reactant used, unimolecular elimination by the conjugate base, intermediate collapse, enzyme-substrate complex cleavage, deamination, intermediate terminated, overall product formed, native state of enzyme regenerated, bimolecular eliminationReferences
- Spraggon G et al. (2001), Proc Natl Acad Sci U S A, 98, 6021-6026. The structures of anthranilate synthase of Serratia marcescens crystallized in the presence of (i) its substrates, chorismate and glutamine, and a product, glutamate, and (ii) its end-product inhibitor, L-tryptophan. DOI:10.1073/pnas.111150298. PMID:11371633.
- Morollo AA et al. (2001), Nat Struct Biol, 8, 243-247. Structure of the cooperative allosteric anthranilate synthase from Salmonella typhimurium. DOI:10.1038/84988. PMID:11224570.
- Knöchel T et al. (1999), Proc Natl Acad Sci U S A, 96, 9479-9484. The crystal structure of anthranilate synthase from Sulfolobus solfataricus: Functional implications. DOI:10.1073/pnas.96.17.9479. PMID:10449718.

Step 1. His175 deprotonates Cys84, activating it for a nucleophilic attack upon L-glutamine, forming an enzyme-substrate covalent bond.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly56B (main-N) | electrostatic stabiliser, hydrogen bond donor |
| Cys84B | hydrogen bond donor |
| His175B | hydrogen bond acceptor, hydrogen bond donor |
| Glu177B | increase basicity, hydrogen bond acceptor |
| Leu85B (main-N) | electrostatic stabiliser, hydrogen bond donor |
| His175B | proton acceptor |
| Cys84B | proton donor, nucleophile |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, enzyme-substrate complex formation, intermediate formation, overall reactant used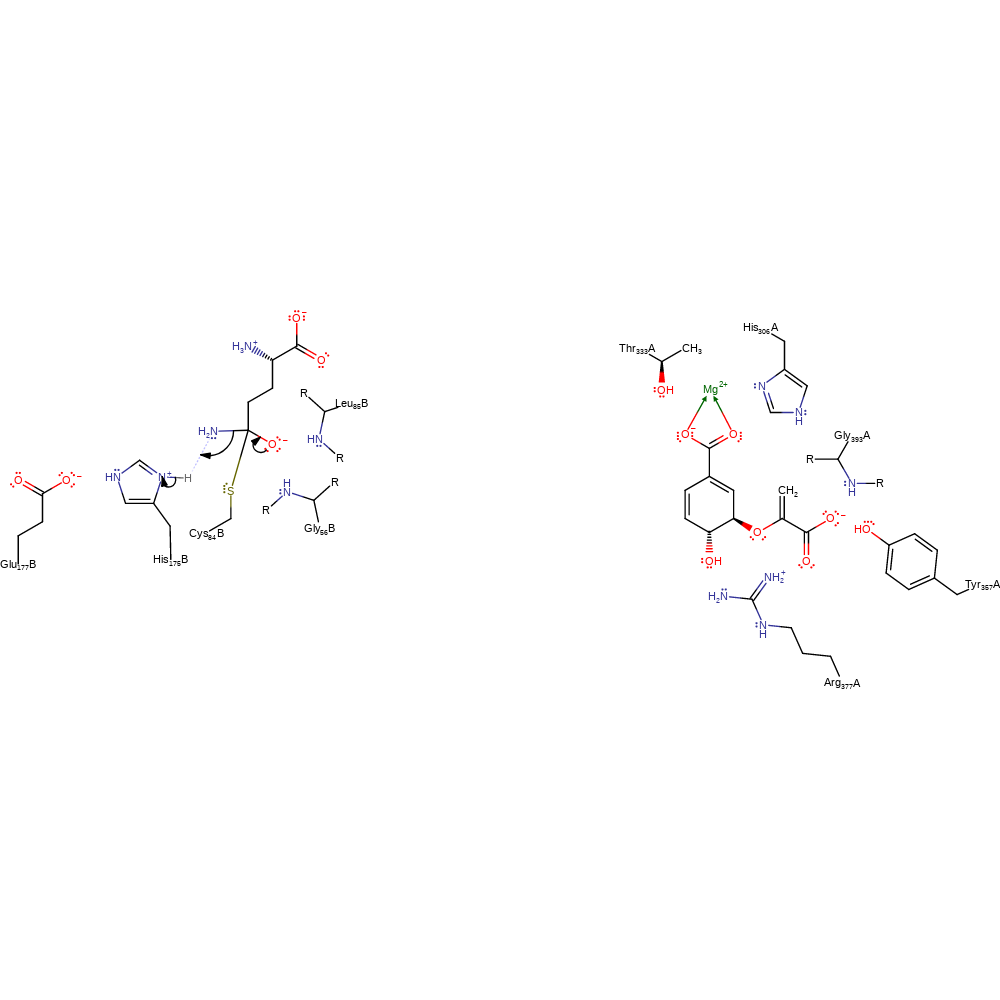
Step 2. The tetrahedral intermediate collapses, liberating ammonia, which deprotonates His175 and then passes to the other catalytic domain.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly56B (main-N) | electrostatic stabiliser, hydrogen bond donor |
| Cys84B | covalently attached |
| His175B | hydrogen bond donor |
| Glu177B | hydrogen bond acceptor, increase acidity |
| Leu85B (main-N) | electrostatic stabiliser, hydrogen bond donor |
| His175B | proton donor |
Chemical Components
ingold: unimolecular elimination by the conjugate base, proton transfer, intermediate collapse, enzyme-substrate complex cleavage, deamination, intermediate formation
Step 3. His175 deprotonates a water molecule, which initiates a nucleophilic attack on the Cys-bound intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly56B (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Cys84B | covalently attached |
| His175B | hydrogen bond donor, hydrogen bond acceptor |
| Glu177B | hydrogen bond acceptor, increase basicity |
| Leu85B (main-N) | hydrogen bond donor, electrostatic stabiliser |
| His175B | proton acceptor |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer, enzyme-substrate complex formation, overall reactant used, intermediate formation
Step 4. The tetrahedral intermediate collapses, liberating Cys84, which deprotonates His175, and the glutamate product.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Gly56B (main-N) | hydrogen bond donor, electrostatic stabiliser |
| Cys84B | covalently attached, hydrogen bond acceptor |
| His175B | hydrogen bond donor |
| Glu177B | hydrogen bond acceptor, increase acidity |
| His175B | proton donor |
| Cys84B | nucleofuge, proton acceptor |
Chemical Components
proton transfer, ingold: unimolecular elimination by the conjugate base, enzyme-substrate complex cleavage, intermediate terminated, intermediate collapse, overall product formed, native state of enzyme regenerated
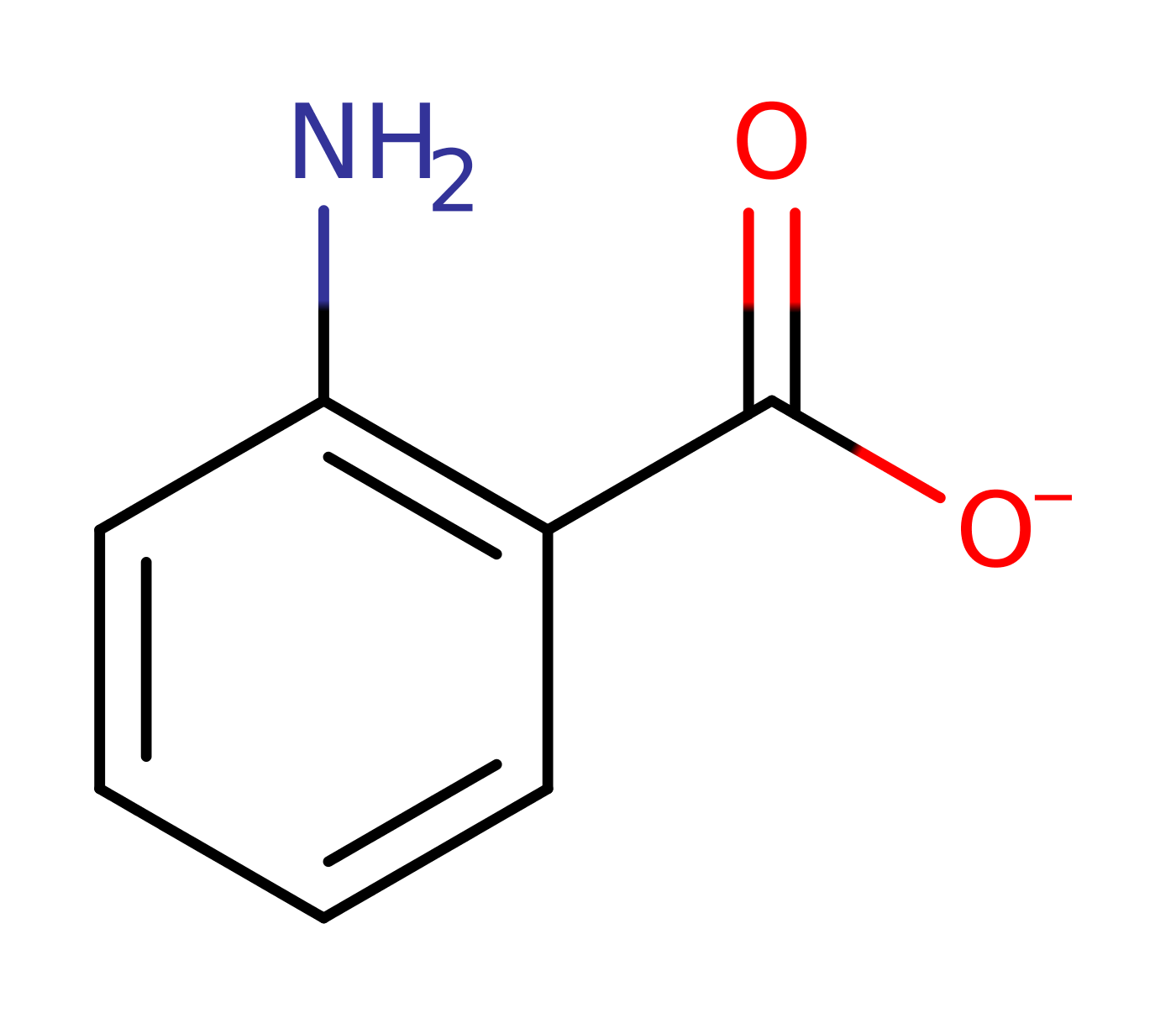
Step 5. Ammiona acts as a nucleophile, adding to chorismate and eliminating water.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr333(334)A | hydrogen bond donor, electrostatic stabiliser |
| His306(307)A | hydrogen bond acceptor |
| Arg377(378)A | hydrogen bond donor, electrostatic stabiliser |
| Tyr357(358)A | hydrogen bond donor, electrostatic stabiliser |
| Gly393(394)A (main-N) | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer, overall reactant used, intermediate formation
Step 6. His306 deprotonates the C2 of the intermediate, eliminating the anthranilate product with concomitant deprotonation of the His306.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr333(334)A | hydrogen bond donor, electrostatic stabiliser |
| His306(307)A | hydrogen bond acceptor, proton relay |
| Arg377(378)A | hydrogen bond donor, electrostatic stabiliser |
| Tyr357(358)A | hydrogen bond donor, electrostatic stabiliser |
| Gly393(394)A (main-N) | hydrogen bond donor, electrostatic stabiliser |
| His306(307)A | proton acceptor, proton donor |





 Download:
Download: