UDP-N-acetylmuramoyl-L-alanine---D-glutamate ligase
UDP-N-acetylmuramoylalanine-D-glutamate ligase catalyses the formation of the peptide bond between UDP-N-acetylmuramoyl-L-alanine and D-glutamate in peptidoglycan biosynthesis. Peptidoglycan is formed as linear repeating disaccharide chains interconnected by a short peptide moiety. It is found in bacterial cell walls and hence peptidoglycan biosynthetic enzymes are possible targets for antibacterial drugs.
Reference Protein and Structure
- Sequence
-
P14900
 (6.3.2.9)
(6.3.2.9)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Escherichia coli K-12 (Bacteria)

- PDB
-
1uag
- UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE
(1.95 Å)



- Catalytic CATH Domains
-
3.40.1190.10
 (see all for 1uag)
(see all for 1uag)
- Cofactors
- Magnesium(2+) (2) Metal MACiE
Enzyme Reaction (EC:6.3.2.9)
Enzyme Mechanism
Introduction
The reaction can be broken down into three steps. The first is the initial phosphorylation of the UMA carboxylate to form the acyl-phosphate. The intermediate would be stabilised by Lys115 and the two divalent cations in the active site. The second is the nucleophilic attack by the amine of D-glutamate producing a tetrahedral intermediate. The formation of the tetrahedral adduct requires a catalytic base to abstract the proton from the D-glutamate amine. This is achieved by either D-glutamate binding in the from solution in the free base form or the deprotonation is assisted by the gamma phosphate of ATP. In the last step the tetrahedral intermediate collapses to yield UDP-N-acetylmuramoyl-L-alanyl-D-glutamate, ADP and inorganic phosphate. This requires another catalytic base to remove the second proton. His183 is thought to act as such a base.
Catalytic Residues Roles
| UniProt | PDB* (1uag) | ||
| His184 | His183A | Acts as a general acid/base. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, steric role |
| Lys116, Asn139 | Lys115A, Asn138A | Acts to stabilise the reactive intermediates and transition states formed during the course of the reaction. | attractive charge-charge interaction, hydrogen bond donor, activator, electrostatic stabiliser, increase electrophilicity |
Chemical Components
bimolecular nucleophilic substitution, overall reactant used, overall product formed, intermediate formation, proton transfer, bimolecular nucleophilic addition, unimolecular elimination by the conjugate base, intermediate collapse, intermediate terminated, inferred reaction step, native state of enzyme regeneratedReferences
- Bertrand JA et al. (1999), J Mol Biol, 289, 579-590. Determination of the MurD mechanism through crystallographic analysis of enzyme complexes. DOI:10.1006/jmbi.1999.2800. PMID:10356330.
- Šink R et al. (2016), PLoS One, 11, e0152075-. Crystallographic Study of Peptidoglycan Biosynthesis Enzyme MurD: Domain Movement Revisited. DOI:10.1371/journal.pone.0152075. PMID:27031227.
- Perdih A et al. (2009), Proteins, 74, 744-759. MurD ligase from E. coli: Tetrahedral intermediate formation study by hybrid quantum mechanical/molecular mechanical replica path method. DOI:10.1002/prot.22188. PMID:18704940.
- Bertrand JA et al. (1997), EMBO J, 16, 3416-3425. Crystal structure of UDP-N-acetylmuramoyl-L-alanine:D-glutamate ligase from Escherichia coli. DOI:10.1093/emboj/16.12.3416. PMID:9218784.
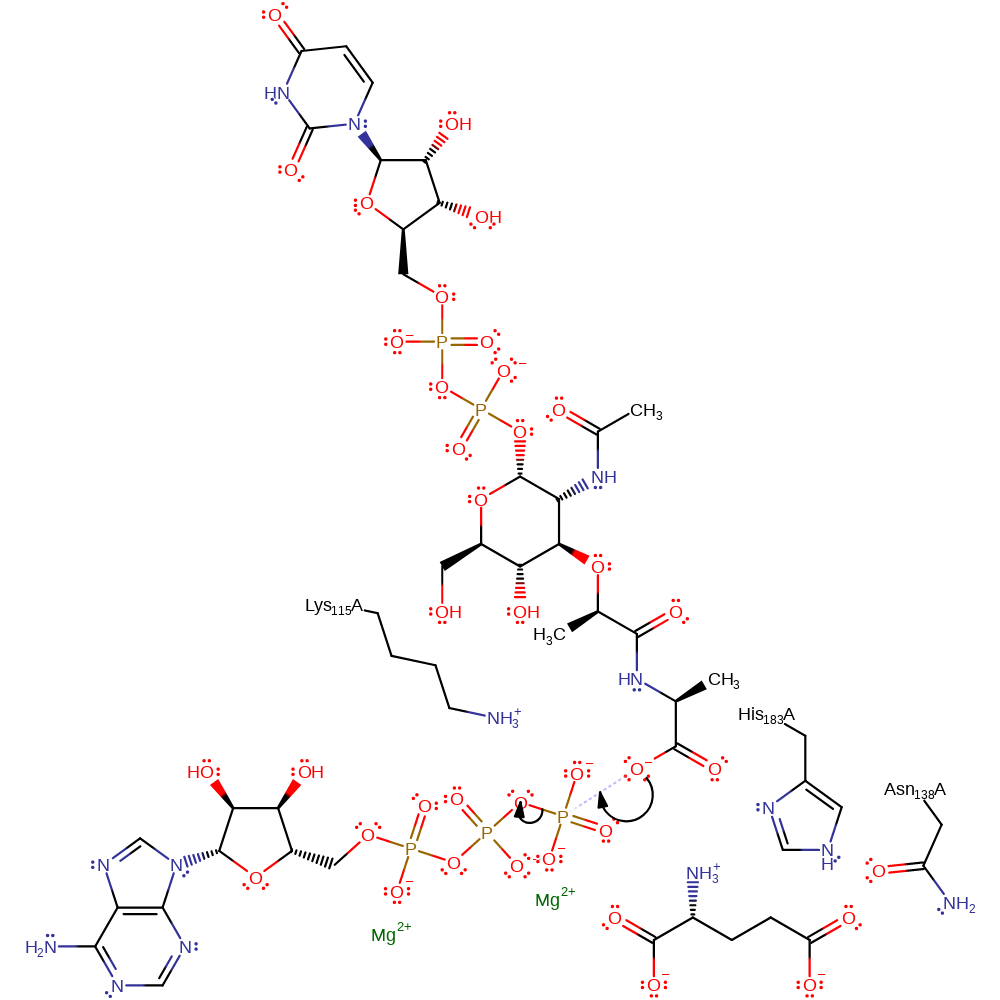
Step 1. The carboxylate group of UDP-N-acetylmuramoyl-L-alanine attacks the gamma phosphate of ATP, forming an activated intermediate and ADP.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys115A | attractive charge-charge interaction, electrostatic stabiliser, increase electrophilicity, activator |
| Asn138A | electrostatic stabiliser, hydrogen bond donor, steric role |
| His183A | hydrogen bond donor, steric role |
Chemical Components
ingold: bimolecular nucleophilic substitution, overall reactant used, overall product formed, intermediate formation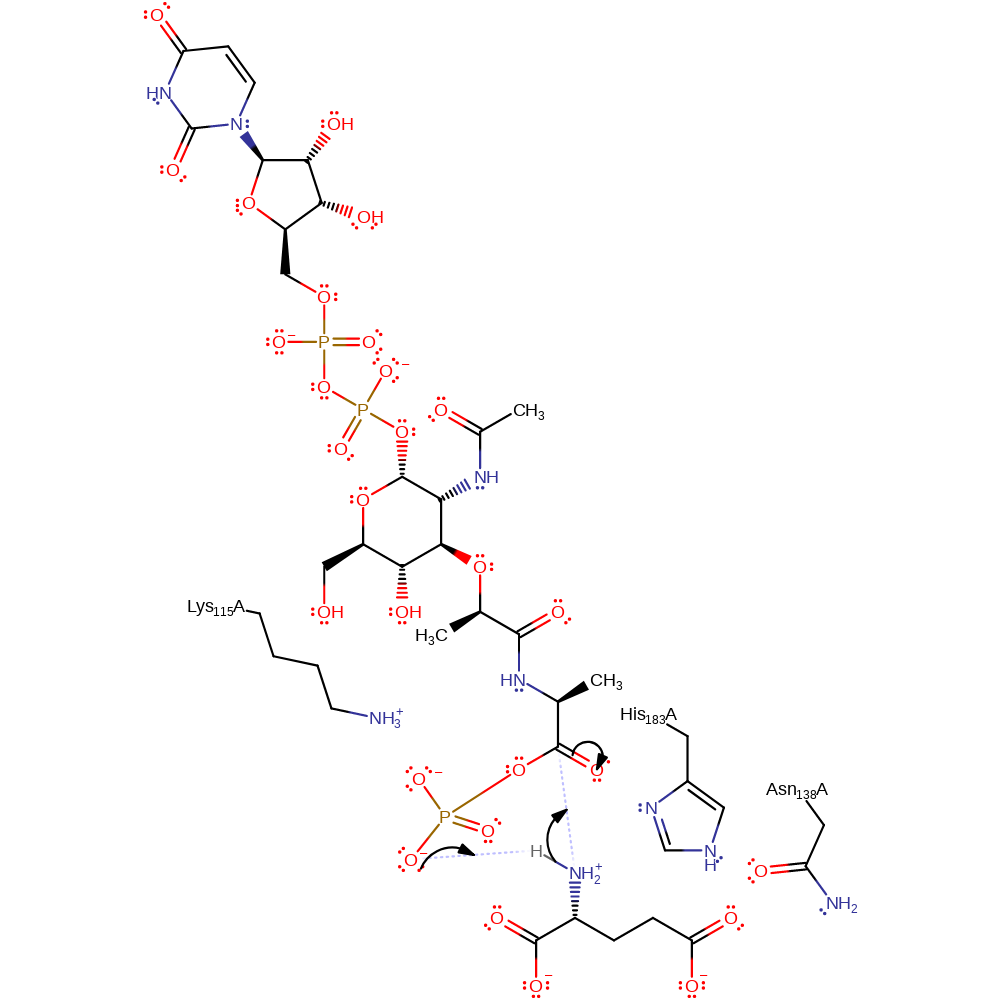
Step 2. The terminal phosphate of the departing ADP abstracts a proton from the amine group of glutamine. The amine group of glutamate then attacks the activated phospho-carbonyl, forming a tetrahedral intermediate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn138A | hydrogen bond donor, steric role, electrostatic stabiliser |
| His183A | hydrogen bond donor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic addition, intermediate formation, overall reactant usedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn138A | hydrogen bond donor, electrostatic stabiliser |
| His183A | hydrogen bond acceptor, hydrogen bond donor |
| His183A | proton acceptor |
Chemical Components
proton transfer, intermediate formation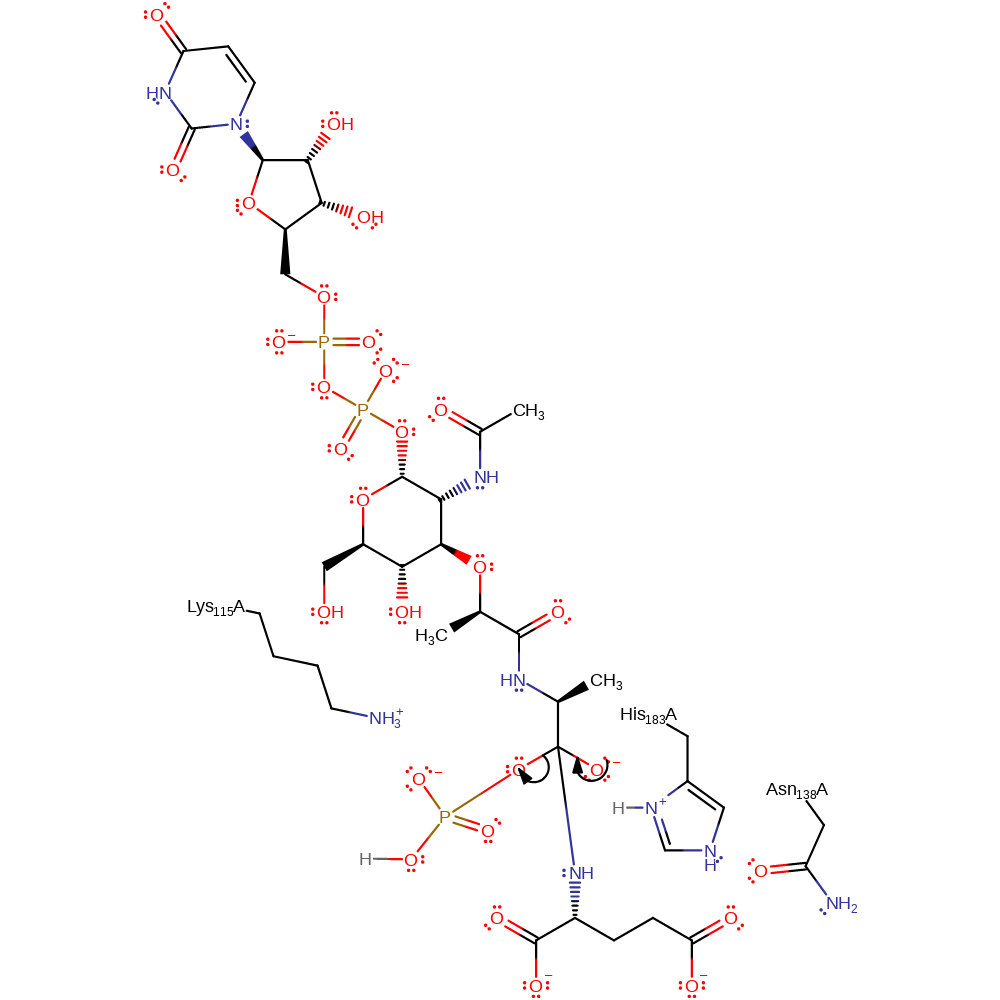
Step 4. The tetrahedral intermediate collapses, releasing a peptidoglycan molecule and protonated phosphate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys115A | hydrogen bond donor |
| Asn138A | electrostatic stabiliser, hydrogen bond donor |
| His183A | hydrogen bond donor |
Chemical Components
ingold: unimolecular elimination by the conjugate base, intermediate collapse, intermediate terminated, overall product formedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His183A | hydrogen bond donor, proton donor |









 Download:
Download: