Glutamate-tRNA ligase
Glutamyl tRNA synthase is able to catalyse the attachment of the amino acid glutamate to its corresponding tRNA molecule, utilising ATP in the process. It is a member of the class I tRNA synthases, and among this group is one of the three known to be unable to catalyse the activation of the amino acid by reaction with ATP unless the tRNA that the amino acid is subsequently transferred to is also present. The enzyme described here, from Thermus thermophilus, displays homology with the other class I tRNA synthases, such as Glutamine tRNA synthase, and is assumed to share a common evolutionary origin with these enzymes, as well as a conserved mechanism by which the reaction occurs.
Reference Protein and Structure
- Sequence
-
P27000
 (6.1.1.17)
(6.1.1.17)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Thermus thermophilus HB8 (Bacteria)

- PDB
-
1j09
- Crystal structure of Thermus thermophilus glutamyl-tRNA synthetase complexed with ATP and Glu
(1.8 Å)



- Catalytic CATH Domains
-
1.10.1160.10
 (see all for 1j09)
(see all for 1j09)
- Cofactors
- Magnesium(2+) (1) Metal MACiE
Enzyme Reaction (EC:6.1.1.17)
Enzyme Mechanism
Introduction
The reaction occurs in two steps, the first reaction of which only occurs in the presence of the bound cognate tRNA molecule. First, in-line nucleophilic attack from the alpha carboxylate oxygen from glutamate on the alpha phosphate of ATP forms glutamylAMP and releases PPi. In this reaction a pentavalent phosphate transition state is formed which is stabilised by the positive charges surrounding the alpha phosphate of ATP, in other words Lys 246 and the Mg2+ ion at the active site. This activates the amino acid by providing it with a good leaving group, so nucleophilic attack from the free 3'OH of the tRNA ribose molecule can occur on the carboxylate, again forming a pentavalent phosphate transition state stabilised by Lys 246 and Mg2+, with the pyrophosphate acting as a base to remove the proton from the 3'OH.
Catalytic Residues Roles
| UniProt | PDB* (1j09) | ||
| Lys246 | Lys246A | Forms contacts with the alpha phosphate of ATP, thus allows stabilisation of the pentavalent phosphate that forms when the amino acid attacks. Also is able to stabilise the second pentavalent phosphate intermediate that forms when the tRNA attacks the glutamyl-AMP intermediate. | attractive charge-charge interaction, increase nucleophilicity, hydrogen bond donor, electrostatic stabiliser |
Chemical Components
bimolecular nucleophilic substitution, intermediate formation, overall reactant used, overall product formed, bimolecular nucleophilic addition, proton transfer, elimination (not covered by the Ingold mechanisms), intermediate collapse, native state of enzyme regeneratedReferences
- Sekine S et al. (2003), EMBO J, 22, 676-688. ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding. DOI:10.1093/emboj/cdg053. PMID:12554668.
- Perona JJ et al. (1993), Biochemistry, 32, 8758-8771. Structural basis for transfer RNA aminoacylation by Escherichia coli glutaminyl-tRNA synthetase. DOI:10.1021/bi00085a006. PMID:8364025.
- Kern D et al. (1980), Eur J Biochem, 106, 137-150. The catalytic mechanism of glutamyl-tRNA synthetase of Escherichia coli. Evidence for a two-step aminoacylation pathway, and study of the reactivity of the intermediate complex. PMID:6280993.
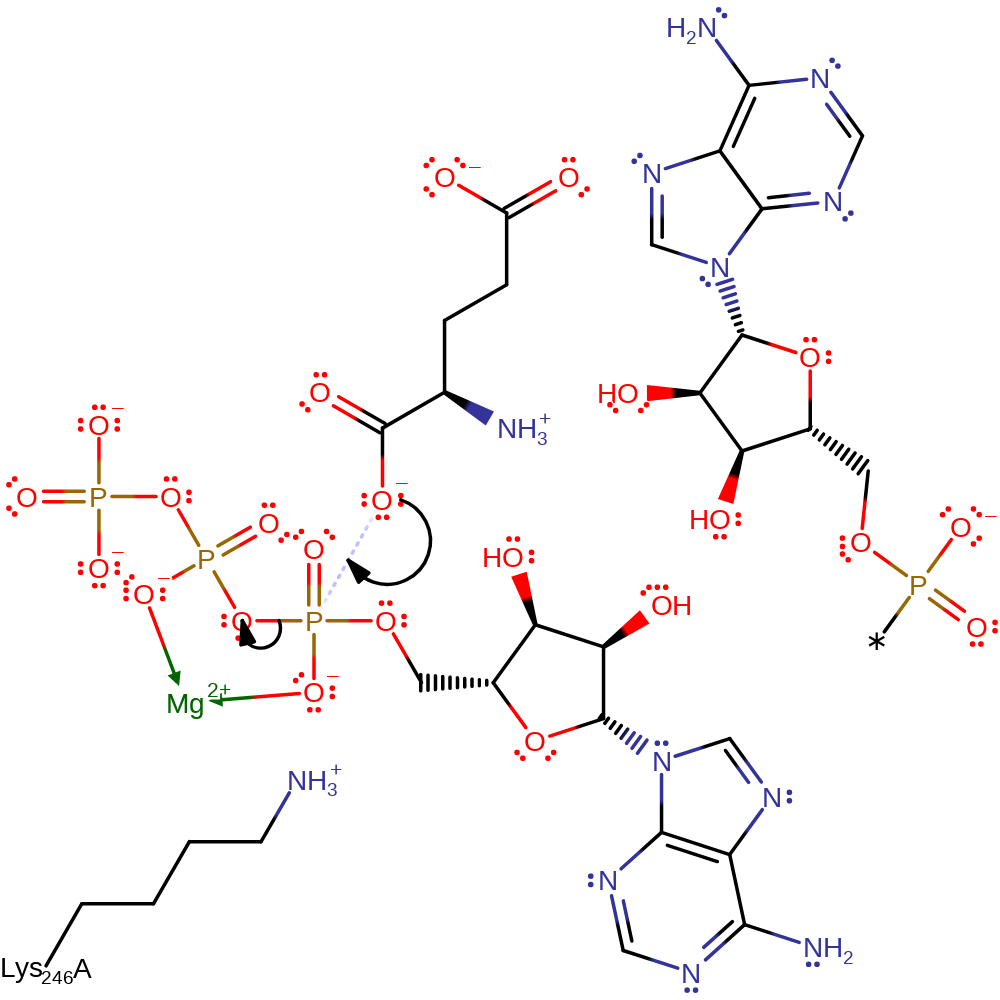
Step 1. The alpha carboxylate group of L-glutamate attacks the alpha phosphate of ATP, forming an activated intermediate and releasing pyrophosphate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys246A | attractive charge-charge interaction, increase nucleophilicity, electrostatic stabiliser |
Chemical Components
ingold: bimolecular nucleophilic substitution, intermediate formation, overall reactant used, overall product formed
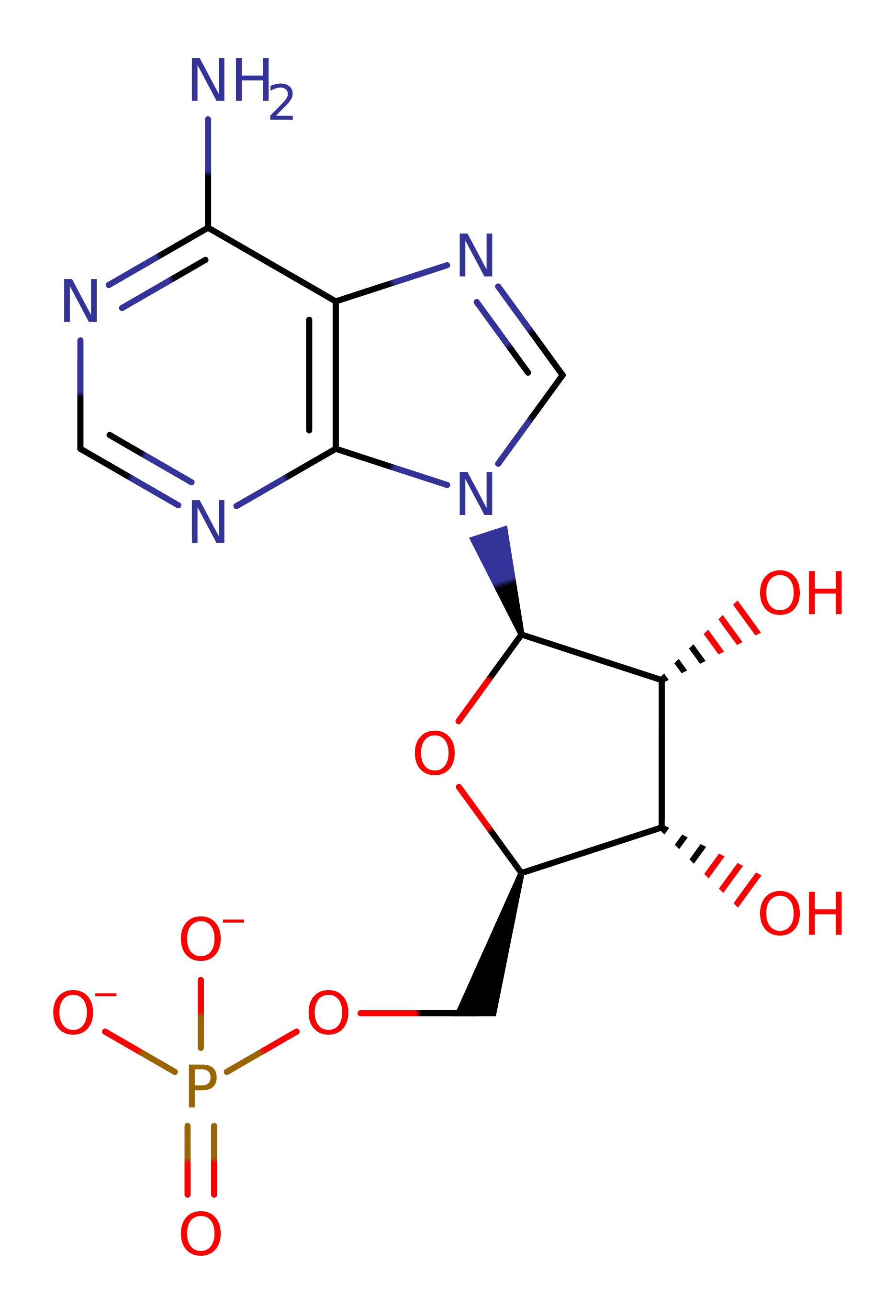
Step 2. The alpha phosphate group of the activated adduct deprotonates the 3'OH of tRNA-Glu, activating the hydroxyl towards attack at the phospho-carbonyl. An anionic tetrahedral intermediate is formed.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys246A | hydrogen bond donor, electrostatic stabiliser |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer, overall reactant used, intermediate formation
Step 3. The anionic intermediate collapses, eliminating AMP and Glu-tRNA.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys246A | hydrogen bond donor, electrostatic stabiliser |




 Download:
Download: