NADPH dehydrogenase
NADPH dehydrogenase, or old yellow enzyme (OYE), isolated from Candida albicans, is a NADPH oxidoreductase. Its FMN cofactor (in plants the cofactor is FAD) is reduced by NADPH, and in turn goes on to reduce the substrate. The exact physiological substrate(s) of OYE are yet to be confirmed, though it is known to act on molecular oxygen, alpha,beta-unsaturated ketones and aldehydes, and quinones. OYE is thought to be involved in general detoxification within the cell.
Reference Protein and Structure
- Sequence
-
Q02899
 (1.6.99.1)
(1.6.99.1)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Saccharomyces pastorianus (Brewer's Yeast)

- PDB
-
1oya
- OLD YELLOW ENZYME AT 2 ANGSTROMS RESOLUTION: OVERALL STRUCTURE, LIGAND BINDING AND COMPARISON WITH RELATED FLAVOPROTEINS
(2.0 Å)



- Catalytic CATH Domains
-
3.20.20.70
 (see all for 1oya)
(see all for 1oya)
- Cofactors
- Fmnh2(2-) (1)
Enzyme Reaction (EC:1.6.99.1)
Enzyme Mechanism
Introduction
This mechanism describes the reduction of an alpha,beta-unsaturated ketone. NADPH transfers a hydride from C4 to the si face of the N5 position of FMN, reducing it and causing a negative charge to build up on N1. Reduced FMN then transfers a hydride from the N5 position to the beta-carbon of the substrate, forming an enolate intermediate that is stabilised by His191, Arg194 and Tyr196. The carbonyl reforms and the alpha-carbon is protonated by Tyr196 to form the saturated carbonyl product.
Catalytic Residues Roles
| UniProt | PDB* (1oya) | ||
| Asn252 | Asn251(252)A | Activates Tyr196 to act as a general acid/base. | hydrogen bond donor, increase acidity |
| His192 | His191(192)A | His191 polarises the carbonyl bond of the substrate, thus making alpha,beta-unsaturated ketone more electrophilic. It also stabilises the enolate intermediate by hydrogen bonding to the negatively charged oxygen. | hydrogen bond donor, electrostatic stabiliser |
| Asn195 | Asn194(195)A | Asn194 polarises the carbonyl bond of the substrate, thus making alpha,beta-unsaturated ketone more electrophilic. It also stabilises the enolate intermediate by hydrogen bonding to the negatively charged oxygen. | hydrogen bond donor, electrostatic stabiliser |
| Thr38 | Thr37(38)A | Thr37 decreases the redox potential of the FMN cofactor by hydrogen bonding to the C4 carbonyl. | hydrogen bond donor, electrostatic stabiliser |
| Tyr197 | Tyr196(197)A | Tyr196 donates a proton to C-alpha of the enolate intermediate to form the saturated ketone product. It is also thought to be able to stabilise the transition state state for the transfer of hydride by interacting with C-alpha. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, electrostatic stabiliser |
Chemical Components
hydride transfer, intermediate formation, overall reactant used, overall product formed, cofactor used, rate-determining step, proton transfer, native state of cofactor regenerated, native state of enzyme regeneratedReferences
- Kohli RM et al. (1998), J Biol Chem, 273, 32763-32770. The Oxidative Half-reaction of Old Yellow Enzyme. THE ROLE OF TYROSINE 196. DOI:10.1074/jbc.273.49.32763. PMID:9830020.
- Brown BJ et al. (2002), J Biol Chem, 277, 2138-2145. The Role of Glutamine 114 in Old Yellow Enzyme. DOI:10.1074/jbc.m108453200. PMID:11668181.
- Xu D et al. (1999), Proc Natl Acad Sci U S A, 96, 3556-3561. The role of threonine 37 in flavin reactivity of the old yellow enzyme. DOI:10.1073/pnas.96.7.3556. PMID:10097075.
- Brown BJ et al. (1998), J Biol Chem, 273, 32753-32762. On the Active Site of Old Yellow Enzyme. ROLE OF HISTIDINE 191 AND ASPARAGINE 194. DOI:10.1074/jbc.273.49.32753. PMID:9830019.
- Fox KM et al. (1994), Structure, 2, 1089-1105. Old yellow enzyme at 2 A resolution: overall structure, ligand binding, and comparison with related flavoproteins. DOI:10.2210/pdb1oyb/pdb. PMID:7881908.
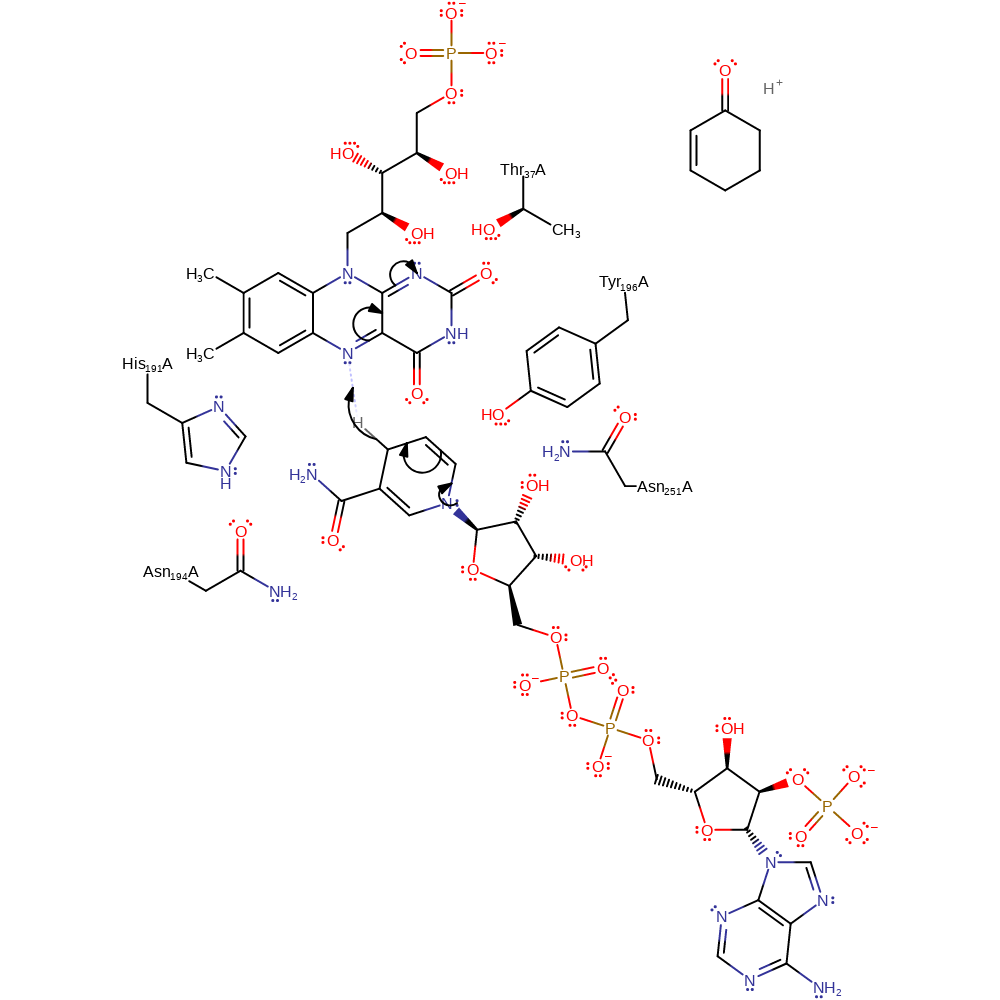
Step 1. NADPH reduces the FMN cofactor. NADPH transfers a hydride from the C4 atom to the si face of the N5 position of FMN [PMID:9830020].
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr37(38)A | hydrogen bond donor, electrostatic stabiliser |
| Tyr196(197)A | electrostatic stabiliser, hydrogen bond donor |
| Asn251(252)A | hydrogen bond donor |
Chemical Components
hydride transfer, intermediate formation, overall reactant used, overall product formed, cofactor used, rate-determining step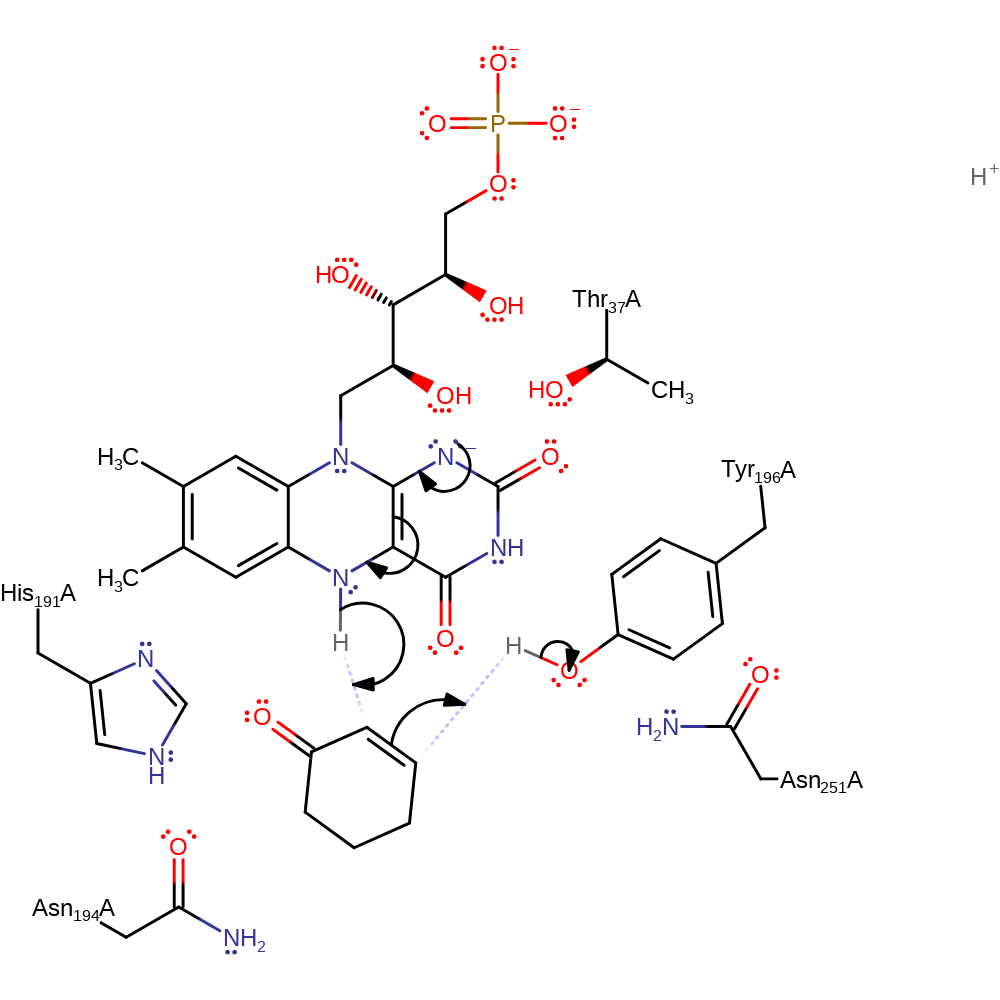
Step 2. The anionic FMN intermediate acts as a hydride donor to the a-b unsaturated substrate. Tyr196 acts as the proton donor in the trans reduction of the substrate double bond. The exact physiological substrate(s) of NADPH dehydrogenase are yet to be confirmed, though it is known to act on molecular oxygen, alpha,beta-unsaturated ketones, aldehydes, and quinones. Reduction across a double bond occurs in a stereospecific trans mechanism [PMID:11668181, PMID:9830020].
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asn194(195)A | hydrogen bond donor, electrostatic stabiliser |
| His191(192)A | hydrogen bond donor, electrostatic stabiliser |
| Thr37(38)A | hydrogen bond donor |
| Tyr196(197)A | hydrogen bond acceptor, hydrogen bond donor |
| Asn251(252)A | increase acidity, hydrogen bond donor |
| Tyr196(197)A | proton donor |
Chemical Components
hydride transfer, proton transfer, native state of cofactor regenerated, overall product formed, overall reactant usedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Tyr196(197)A | hydrogen bond donor, hydrogen bond acceptor |
| Thr37(38)A | hydrogen bond donor |
| Asn251(252)A | hydrogen bond donor |
| Tyr196(197)A | proton acceptor |




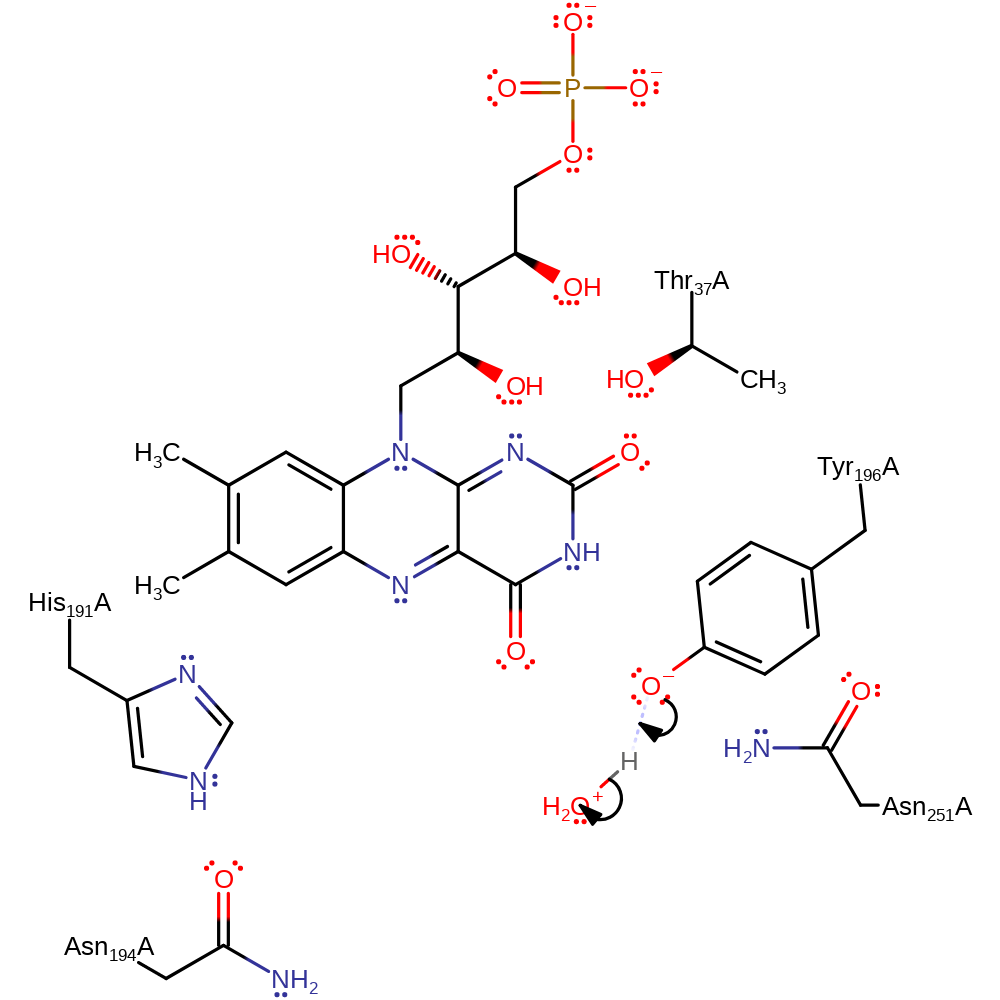
 Download:
Download: