Sulfide:quinone oxidoreductase
Catalyses the oxidation of sulfides, such as hydrogen sulfide, with the help of a quinone to form sulfur globules. Consecutive reaction cycles lead to the accumulation of a polysulfide product on the active site Cys residues; these products are released when they exceed a critical length, typically as cyclooctasulfur. This is an important step in anoxygenic bacterial photosynthesis.
Reference Protein and Structure
- Sequence
-
Q7ZAG8
 (1.8.5.4)
(1.8.5.4)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Acidianus ambivalens (Archaea)

- PDB
-
3h8i
- The first X-ray structure of a sulfide:quinone oxidoreductase: Insights into sulfide oxidation mechanism
(2.65 Å)



- Catalytic CATH Domains
-
3.50.50.60
 (see all for 3h8i)
(see all for 3h8i)
- Cofactors
- Fadh2(2-) (1)
Enzyme Reaction (EC:1.8.5.4)
Enzyme Mechanism
Introduction
This enzyme has the highest activity with caldariella quinone and decylubiquinone, and lower activity with naphtoquinones. The substrate hydrogen sulfide is deprotonated by Asp353, activating the sulfur as a nucleophile towards the protein disulfide bridge. The anionic Cys178A attacks the C4 position of FAD, initiating protonation of the N5 position. The Cys350 dithiolate acts as a nucleophile towards the enzyme cofactor adduct, forming a three centre sulfur intermediate and the reduced form of FAD. Ubiquinone, the physiological electron acceptor accepts a hydride from reduced FAD, regenerating the cofactor for the next round of sulfide oxidiation. Only the first four steps are shown, but this enzyme catalyses several successive rounds of catalysis, until the polysulfide product on the active site Cys residues exceeds a critical length.
Catalytic Residues Roles
| UniProt | PDB* (3h8i) | ||
| Cys178, Cys350 | Cys178A, Cys350A | In the ground state of the protein, Cys350 and Cys178 are covalently attached in a disulfide bond. During the course of the reaction both residues act via covalent catalysis. Cys178 becomes attached to the FAD cofactor and is finally added back onto the Cys350, which during the course of the reaction accepts the substrate sulfur atom. | covalently attached, nucleofuge, nucleophile, activator, electrofuge, electrophile |
| Asp215, Asp353 | Asp215A, Asp353A | Forms a proton relay chain that ultimately deprotonates the substrate hydrogen sulfide; both residues act as a general acid/base. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, activator |
| Cys129 | Cys129A | Forms a covalent bond with the FAD cofactor, enhancing its redox potential. | covalently attached, activator |
| Lys386 | Lys386A | Helps stabilise the negative charge in the active site. This residue possibly acts as a general acid/base in the regeneration of the FAD cofactor stage of the reaction. | activator |
Chemical Components
proton transfer, bimolecular nucleophilic substitution, overall reactant used, enzyme-substrate complex formation, bimolecular nucleophilic addition, cofactor used, intermediate formation, enzyme-substrate complex cleavage, hydride transfer, overall product formed, native state of cofactor regeneratedReferences
- Brito JA et al. (2009), Biochemistry, 48, 5613-5622. Structural and Functional Insights into Sulfide:Quinone Oxidoreductase,. DOI:10.1021/bi9003827. PMID:19438211.
- Cherney MM et al. (2010), J Mol Biol, 398, 292-305. Crystal Structure of Sulfide:Quinone Oxidoreductase from Acidithiobacillus ferrooxidans: Insights into Sulfidotrophic Respiration and Detoxification. DOI:10.1016/j.jmb.2010.03.018. PMID:20303979.
- Marcia M et al. (2009), Proc Natl Acad Sci U S A, 106, 9625-9630. The structure of Aquifex aeolicus sulfide:quinone oxidoreductase, a basis to understand sulfide detoxification and respiration. DOI:10.1073/pnas.0904165106. PMID:19487671.

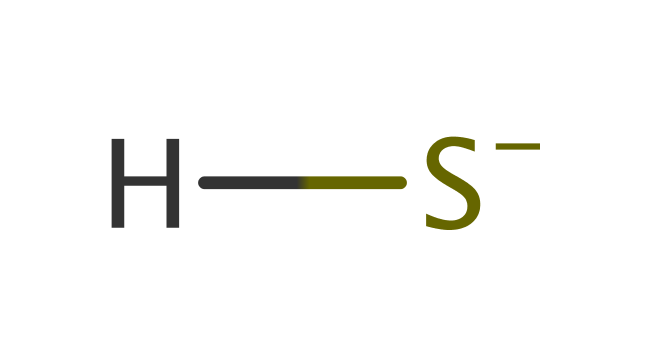
Step 1. The substrate hydrogen sulfide is deprotonated by Asp353, activating the sulfur as a nucleophile towards the protein disulfide bridge. The sulfide oxidation occurs on the re face of the FAD cofactor [PMID:19438211].
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp353A | activator, hydrogen bond acceptor |
| Asp215A | activator |
| Cys350A | activator |
| Cys178A | activator |
| Cys129A | covalently attached |
| Asp353A | proton donor, proton acceptor |
| Cys350A | electrophile |
| Cys178A | nucleofuge |
| Cys350A | electrofuge |
| Asp215A | proton acceptor |
| Asp353A | proton relay |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic substitution, overall reactant used, enzyme-substrate complex formation
Step 2. The anionic Cys178A attacks the C4 position of FAD, initiating protonation of the N5 position.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp215A | hydrogen bond donor |
| Cys350A | activator, covalently attached |
| Cys178A | activator |
| Asp353A | hydrogen bond donor, hydrogen bond acceptor |
| Cys129A | covalently attached |
| Cys178A | nucleophile |
Chemical Components
ingold: bimolecular nucleophilic addition, proton transfer, cofactor used, intermediate formation
Step 3. The Cys350 dithiolate acts as a nucleophile towards the enzyme cofactor adduct, forming a three centre sulfur intermediate and the reduced form of FAD. The steric bulk brought about by the presence of an elongated sulfur chain attached Cys350A directs the anionic Cys178A towards nucleophilic attack at Cys350A, reforming the active site disulfide bridge.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp215A | activator |
| Cys350A | activator, covalently attached |
| Cys178A | activator, covalently attached |
| Asp353A | activator, hydrogen bond acceptor, hydrogen bond donor |
| Cys129A | activator, covalently attached |
| Asp353A | proton acceptor |
| Asp215A | proton donor |
| Asp353A | proton relay |
| Cys178A | electrofuge |
| Asp353A | proton donor |
| Cys178A | electrophile |
Chemical Components
ingold: bimolecular nucleophilic substitution, proton transfer, cofactor used, enzyme-substrate complex cleavage, intermediate formation
Step 4. Ubiquinone, the physiological electron acceptor accepts a hydride from reduced FAD, regenerating the cofactor for the next round of sulfide oxidiation. While potential proton donors to the ubiquinone identified in Acidithiobacillus ferrooxidans are conserved in the Acidianus ambivalens homolouge listed here, no general base to the FAD cofactor has been suggested in either case [PMID:20303979].
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp215A | hydrogen bond acceptor |
| Cys350A | covalently attached |
| Cys178A | covalently attached |
| Asp353A | hydrogen bond donor |
| Lys386A | activator |
| Cys129A | covalently attached |




 Download:
Download: