Pyruvate kinase
Pyruvate kinase catalyses the final step of glycolysis and is allosterically regulated by fructose-1,6-bisphosphate. It requires two equivalents divalent cation, one of which binds to the enzyme as a complex with the nucleotide substrate, as well as activation by K+. As opposed to mammalian pyruvate kinase, the bacterial enzyme works in a K+ independent manner.
The enzyme is also known to catalyse a variety of side reactions, including the decarboxylation of oxalacetate, the enolisation of pyruvate, ATP-dependent phosphorylation of alpha-hydroxy or alpha-thio carboxylates, ATP- and bicarbonate-dependent phosphorylations of fluoride and of hydroxylamine and to also function as an ATP- and bicarbonate-dependent ATPase. These side activities reflect the capacity of the active site to labilise the gamma-phosphate of ATP or to stabilise the enolate of pyruvate [PMID:9308890].
There are four isozymes of pyruvate kinase in mammals (L, R, M1, M2) encoded by two different genes: PKLR and PKM. The L and R isozymes are generated from the PKLR by differential splicing of RNA; the M1 and M2 forms are produced from the PKM gene by differential splicing. L type is major isozyme in the liver, R is found in red cells, M1 is the main form in muscle, heart and brain, and M2 is found in early fetal tissues as well as in most cancer cells. This entry represents the PKM gene products.
Reference Protein and Structure
- Sequence
-
P11974
 (2.7.1.40)
(2.7.1.40)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Oryctolagus cuniculus (rabbit)

- PDB
-
1pkn
- STRUCTURE OF RABBIT MUSCLE PYRUVATE KINASE COMPLEXED WITH MN2+, K+, AND PYRUVATE
(2.9 Å)



- Catalytic CATH Domains
-
3.20.20.60
 2.40.33.10
2.40.33.10  (see all for 1pkn)
(see all for 1pkn)
- Cofactors
- Potassium(1+) (1), Manganese(2+) (1) Metal MACiE
Enzyme Reaction (EC:2.7.1.40)
Enzyme Mechanism
Introduction
The reaction of pyruvate kinase is a two step reaction. In the first reaction the phosphate group of PEP is transferred to ADP to produce ATP. The bound enol is then protonated to produce pyruvate in its keto form.
Catalytic Residues Roles
| UniProt | PDB* (1pkn) | ||
| Lys270 | Lys269A | Stabilises the transition state, also acts to bring the substrates together in the correct orientation for the reaction to occur. Also interacts with the potassium ion (although not a ligand). | attractive charge-charge interaction, hydrogen bond donor, electrostatic stabiliser, steric role |
| Thr328 | Thr327A | The Thr237 residue has been shown to influence the pKa of the specific acid, and is positioned within the active site to influence the stereo-selectivity of the protonation, although kinetic and pH studies have shown the proton to originate from a solvent molecule [PMID:15568816]. | hydrogen bond donor, electrostatic stabiliser, increase acidity |
| Arg73, Arg120 | Arg72A, Arg119A | Stabilises the transition state. Also interacts with the potassium ion (although not a ligand). | attractive charge-charge interaction, electrostatic stabiliser, steric role |
Chemical Components
bimolecular nucleophilic substitution, overall reactant used, overall product formed, intermediate formation, dephosphorylation, proton transfer, intermediate collapse, native state of enzyme regeneratedReferences
- Larsen TM et al. (1997), Arch Biochem Biophys, 345, 199-206. Ligand-Induced Domain Movement in Pyruvate Kinase: Structure of the Enzyme from Rabbit Muscle with Mg2+, K+, andl-Phospholactate at 2.7 Å Resolution. DOI:10.1006/abbi.1997.0257. PMID:9308890.
- Israelsen WJ et al. (2015), Semin Cell Dev Biol, 43, 43-51. Pyruvate kinase: Function, regulation and role in cancer. DOI:10.1016/j.semcdb.2015.08.004. PMID:26277545.
- Ramírez-Silva L et al. (2014), Int J Mol Sci, 15, 22214-22226. The importance of polarity in the evolution of the K+ binding site of pyruvate kinase. DOI:10.3390/ijms151222214. PMID:25474090.
- Susan-Resiga D et al. (2004), Biochemistry, 43, 15230-15245. Proton Donor in Yeast Pyruvate Kinase: Chemical and Kinetic Properties of the Active Site Thr 298 to Cys Mutant†. DOI:10.1021/bi049864d. PMID:15568816.
- Susan-Resiga D et al. (2003), J Biol Chem, 278, 12660-12671. The Proton Transfer Step Catalyzed by Yeast Pyruvate Kinase. DOI:10.1074/jbc.m300257200. PMID:12562754.
- Bollenbach TJ et al. (1999), Biochemistry, 38, 9137-9145. Role of Lysine 240 in the Mechanism of Yeast Pyruvate Kinase Catalysis†. DOI:10.1021/bi990690n. PMID:10413488.
- Larsen TM et al. (1994), Biochemistry, 33, 6301-6309. Structure of rabbit muscle pyruvate kinase complexed with Mn2+, K+, and pyruvate. DOI:10.2210/pdb1pkn/pdb. PMID:8193145.

Step 1. Phosphoryl transfer from PEP to M(II)ADP occurs by an apparent SN2 mechanism with an inversion of configuration at the phosphoryl group, to yield the enolate of pyruvate and M(II)ATP. The presence of either a Mn or Mg divalent cation is essential for catalytic activity [PMID:8193145]. The monovalent cation, K+, does not directly contact the substrate or intermediate, but instead is thought to influence the structure of the active site though interactions with the positively charged resides Arg72, Arg119 and Lys269 [PMID:8193145].
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg72A | attractive charge-charge interaction, electrostatic stabiliser |
| Lys269A | attractive charge-charge interaction, hydrogen bond donor, electrostatic stabiliser, steric role |
| Arg119A | attractive charge-charge interaction, electrostatic stabiliser |
| Thr327A | hydrogen bond donor |
Chemical Components
ingold: bimolecular nucleophilic substitution, overall reactant used, overall product formed, intermediate formation, dephosphorylation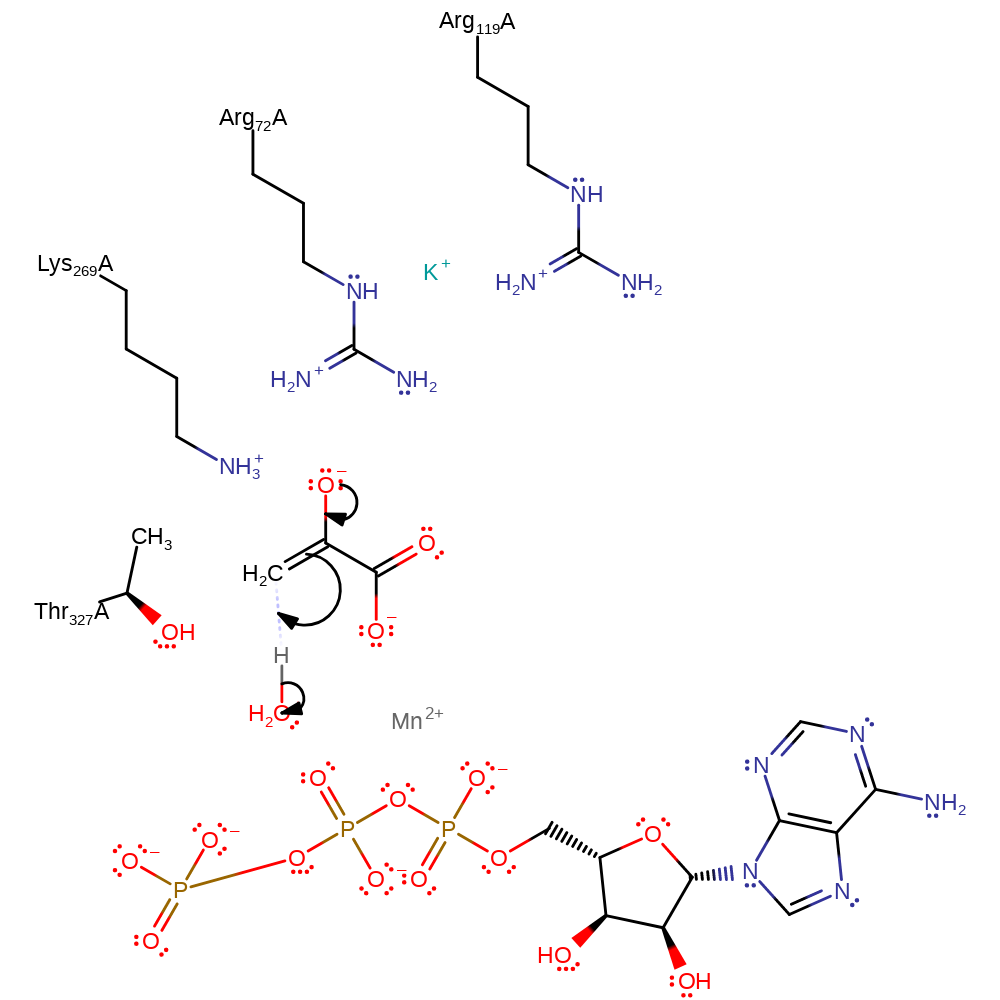
Step 2. Thr237 and the Mn(II) cation increase the acidity of water through hydrogen bonding and charge stabilisation, respectively, generating a specific acid to act towards the enolate intermediate. A solvent molecule at the active site stereospecifically protonates the enolate at the 2-si face of the double bond to form keto pyruvate. Kinetic and pH studies have shown the proton to originate from a solvent molecule [PMID:15568816]. The divalent cation is thought to enhance the acidity of the solvent molecule, rather than the monovalent cation [PMID:12562754].
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Arg72A | attractive charge-charge interaction, electrostatic stabiliser, steric role |
| Lys269A | attractive charge-charge interaction, electrostatic stabiliser, steric role |
| Arg119A | attractive charge-charge interaction, electrostatic stabiliser, steric role |
| Thr327A | hydrogen bond donor, increase acidity, electrostatic stabiliser |





 Download:
Download: