Inositol-3-phosphate synthase
Inositol-3-phosphate synthase catalyses the conversion of D-glucose 6-phosphate to 1L-myo-inositol-1-phosphate, the first committed step in the production of all inositol-containing compounds, including phospholipids, either directly or by salvage. It requires NAD+, which dehydrogenates the -CHOH- group to -CO- at C-5 of the glucose 6-phosphate, making C-6 into an active methylene, able to condense with the -CHO at C-1. Finally, the enzyme-bound NADH reconverts C-5 into the -CHOH- form.
Reference Protein and Structure
- Sequence
-
P11986
 (5.5.1.4)
(5.5.1.4)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Saccharomyces cerevisiae S288c (Baker's yeast)

- PDB
-
1rm0
- Crystal Structure of Myo-Inositol 1-Phosphate Synthase From Saccharomyces cerevisiae In Complex With NAD+ and 2-deoxy-D-glucitol 6-(E)-vinylhomophosphonate
(2.05 Å)



- Catalytic CATH Domains
-
3.30.360.10
 3.40.50.720
3.40.50.720  (see all for 1rm0)
(see all for 1rm0)
- Cofactors
- Nadh(2-) (1)
Enzyme Reaction (EC:5.5.1.4)
Enzyme Mechanism
Introduction
The substrate is oxidised at the C5' by NAD+, with direct hydride transfer from C5 to C4 of the nicotinamide ring in concert with proton loss from the C5' hydroxyl group of D-glucose-6-phosphate. This proton is transferred to the Lys369 side chain. The pro-R hydrogen of C6 is eliminated. Evidence from crystallographic data suggests the phosphate-mono ester acts as the base in the enolisation step. The developing negative charge on the enolate oxygen is stabilised by two lysine residues, Lys369 and Lys489. In this aldol condensation step either the monophosphate or lys412 acts as a general acid to O1 in the cyclisation step. The last mechanistic step is reduction by NADPH. The hydride that was transferred in the first step is returned to the C5 position of the intermediate myo-2-inose 1-phosphate. The active site is regenerated on protonation.
Catalytic Residues Roles
| UniProt | PDB* (1rm0) | ||
| Lys412 | Lys412A | Acts as a general acid/base involved in ring opening and closing. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, activator, electrostatic stabiliser |
| Lys489, Lys369 | Lys489A, Lys369A | Acts as a general acid/base. Also helps to stabilise the negative charge on the enolate oxygen. | proton acceptor, hydrogen bond donor, electrostatic stabiliser, proton donor |
| Asp320 | Asp320A | Important for the stabilisation of the resulting positive charge by acting as a hydrogen bond acceptor to the general acid/base Lys369. | hydrogen bond acceptor, electrostatic stabiliser |
Chemical Components
hydride transfer, proton transfer, intermediate formation, overall reactant used, cofactor used, assisted keto-enol tautomerisation, intramolecular nucleophilic addition, overall product formed, native state of cofactor regenerated, native state of enzyme regenerated, inferred reaction stepReferences
- Jin X et al. (2004), J Biol Chem, 279, 13889-13895. The Structure of the 1L-myo-inositol-1-phosphate Synthase-NAD+-2-deoxy-D-glucitol 6-(E)-Vinylhomophosphonate Complex Demands a Revision of the Enzyme Mechanism. DOI:10.1074/jbc.m308986200. PMID:14684747.
- Seelan RS et al. (2009), J Biol Chem, 284, 9443-9457. Identification of myo-Inositol-3-phosphate Synthase Isoforms: CHARACTERIZATION, EXPRESSION, AND PUTATIVE ROLE OF A 16-kDa c ISOFORM. DOI:10.1074/jbc.m900206200. PMID:19188364.
- Stein AJ et al. (2002), J Biol Chem, 277, 9484-9491. The Crystal Structure and Mechanism of 1-L-myo-Inositol- 1-phosphate Synthase. DOI:10.1074/jbc.m109371200. PMID:11779862.

Step 1. The substrate is oxidised at the C5' by NAD+, with direct hydride transfer from C5 to C4 of the nicotinamide ring in concert with proton loss from the C5' hydroxyl group of D-glucose-6-phosphate. This proton is transferred to the Lys369 side chain.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys412A | hydrogen bond donor |
| Asp320A | electrostatic stabiliser, hydrogen bond acceptor |
| Lys369A | activator, hydrogen bond acceptor |
| Lys489A | hydrogen bond donor, electrostatic stabiliser |
| Lys369A | proton acceptor |
Chemical Components
hydride transfer, proton transfer, intermediate formation, overall reactant used, cofactor used
Step 2. The pro-R hydrogen of C6 is eliminated. Evidence from crystallographic data suggests the phosphate-mono ester acts as the base in the enolisation step. The developing negative charge on the enolate oxygen is stabilised by two lysine residues, Lys369 and Lys489.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys412A | hydrogen bond donor |
| Asp320A | electrostatic stabiliser, hydrogen bond acceptor |
| Lys369A | electrostatic stabiliser, hydrogen bond donor |
| Lys489A | electrostatic stabiliser, hydrogen bond donor |
Chemical Components
assisted keto-enol tautomerisation, intermediate formation
Step 3. In this aldol condensation step either the monophosphate or Lys412 acts as a general acid to O1 in the cyclisation step. The side chain of Lys373 is thought to aid in stabilising the O1 negative charge [PMID:14684747].
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys412A | activator, electrostatic stabiliser, hydrogen bond donor |
| Asp320A | electrostatic stabiliser, hydrogen bond acceptor |
| Lys369A | electrostatic stabiliser, hydrogen bond donor |
| Lys489A | electrostatic stabiliser, hydrogen bond donor |
| Lys412A | proton donor |
Chemical Components
assisted keto-enol tautomerisation, ingold: intramolecular nucleophilic addition, proton transfer, intermediate formation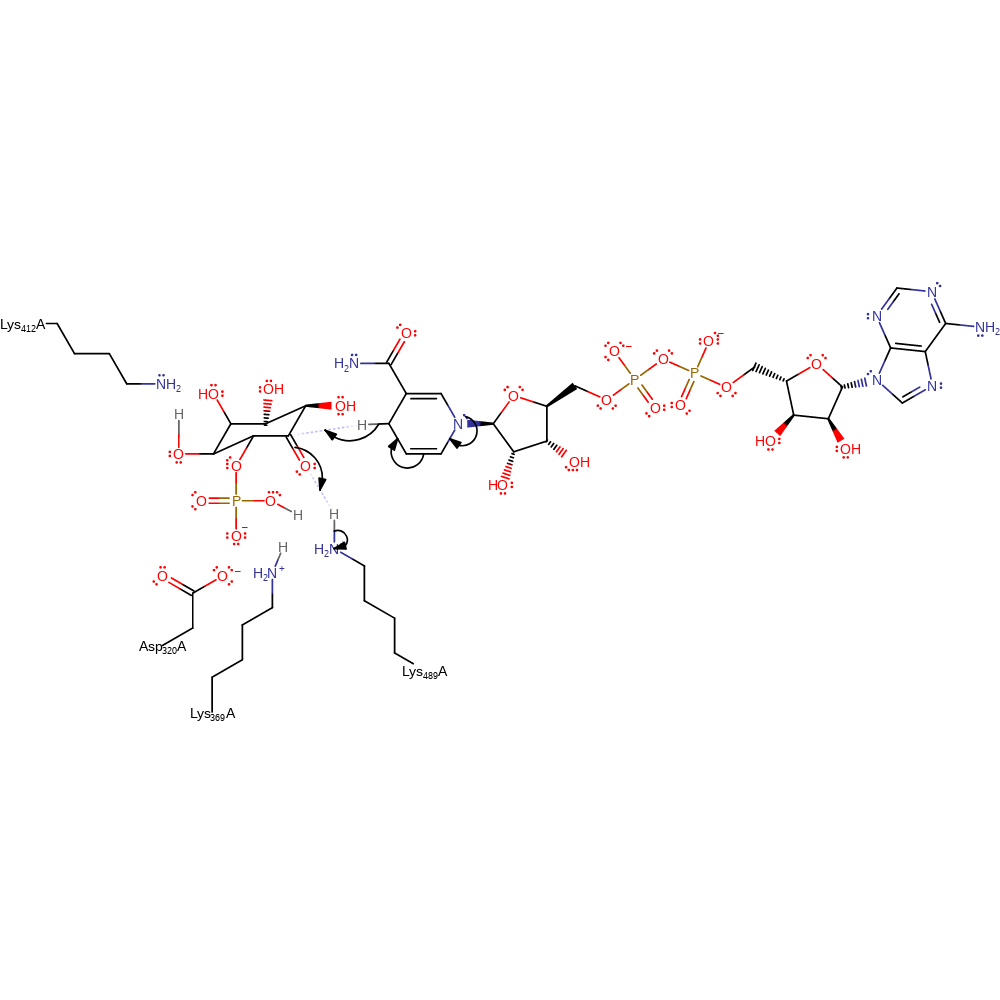
Step 4. The last mechanistic step is reduction by NADPH. The hydride that was transferred in the first step is returned to the C5 position of the intermediate myo-2-inose 1-phosphate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys412A | hydrogen bond acceptor |
| Asp320A | electrostatic stabiliser, hydrogen bond acceptor |
| Lys369A | activator, hydrogen bond donor |
| Lys489A | electrostatic stabiliser, hydrogen bond donor |
| Lys489A | proton donor |
Chemical Components
hydride transfer, proton transfer, overall product formed, native state of cofactor regenerated, cofactor usedCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Lys412A | activator, hydrogen bond acceptor |
| Asp320A | hydrogen bond acceptor |
| Lys369A | hydrogen bond donor |
| Lys489A | hydrogen bond donor, proton acceptor |
| Lys412A | proton acceptor |
| Lys369A | proton donor |



 Download:
Download: