1-deoxy-D-xylulose 5-phosphate synthase
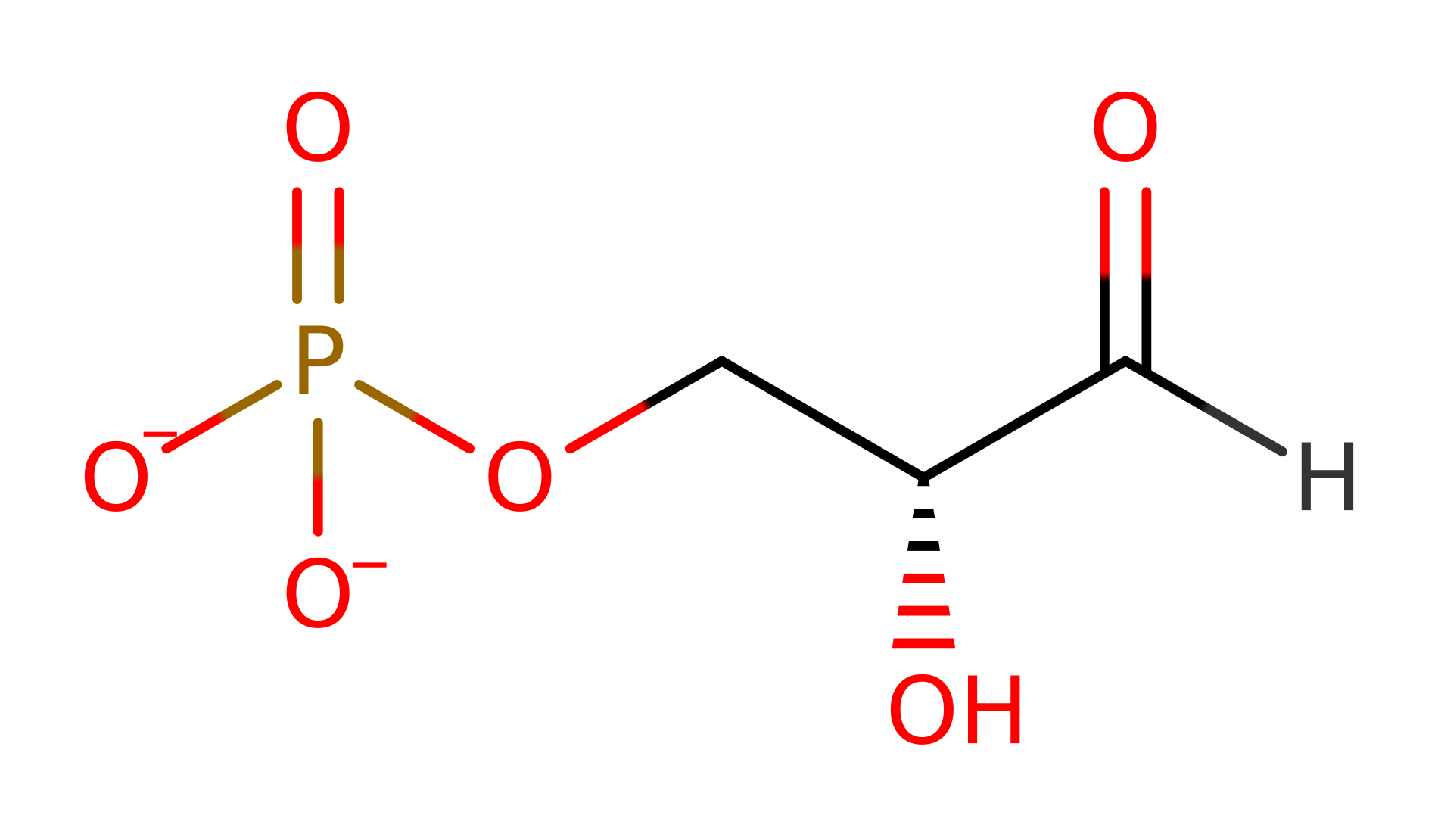
1-Deoxy-D-xylulose-5-phosphate synthase (DXS ) is a regulatory enzyme of the mevalonate-independent pathway involved in terpenoid biosynthesis. DXP synthase is a thiamine diphosphate-dependent enzyme related to transketolase and the pyruvate dehydrogenase E1-beta subunit. DXS is found in bacteria (gene:dxs) and plants (gene:CLA1) which catalyses the thiamine pyrophosphoate-dependent acyloin condensation reaction between carbon atoms 2 and 3 of pyruvate and glyceraldehyde 3-phosphate to yield 1-deoxy-D-xylulose-5-phosphate (DXP), a precursor in the biosynthetic pathway to isoprenoids, thiamine (vitamin B1), and pyridoxol (vitamin B6). DXS is evolutionary related to TK. This reaction is the first and rate limiting step of the mevalonate-independent pathway for the biosynthesis of isopentyl pyrophosphate, a process undertaken only in fungi, algae and bacteria. DXS differs from related thiamine diphosphate dependent enzymes by virtue of its domain arrangement: the active site is not situated on a chain interface, as seen in related enzymes, but instead resides between two domain faces within the same monomer of a homodimeric complex [PMID:17135236].
Reference Protein and Structure
- Sequence
-
Q9RUB5
 (2.2.1.7)
(2.2.1.7)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Deinococcus radiodurans R1 (Bacteria)

- PDB
-
2o1x
- 1-deoxy-D-xylulose 5-phosphate synthase (DXS) from Deinococcus radiodurans
(2.9 Å)



- Catalytic CATH Domains
-
3.40.50.970
 (see all for 2o1x)
(see all for 2o1x)
- Cofactors
- Thiamine(1+) diphosphate(3-) (1), Magnesium(2+) (1)
Enzyme Reaction (EC:2.2.1.7)
Enzyme Mechanism
Introduction
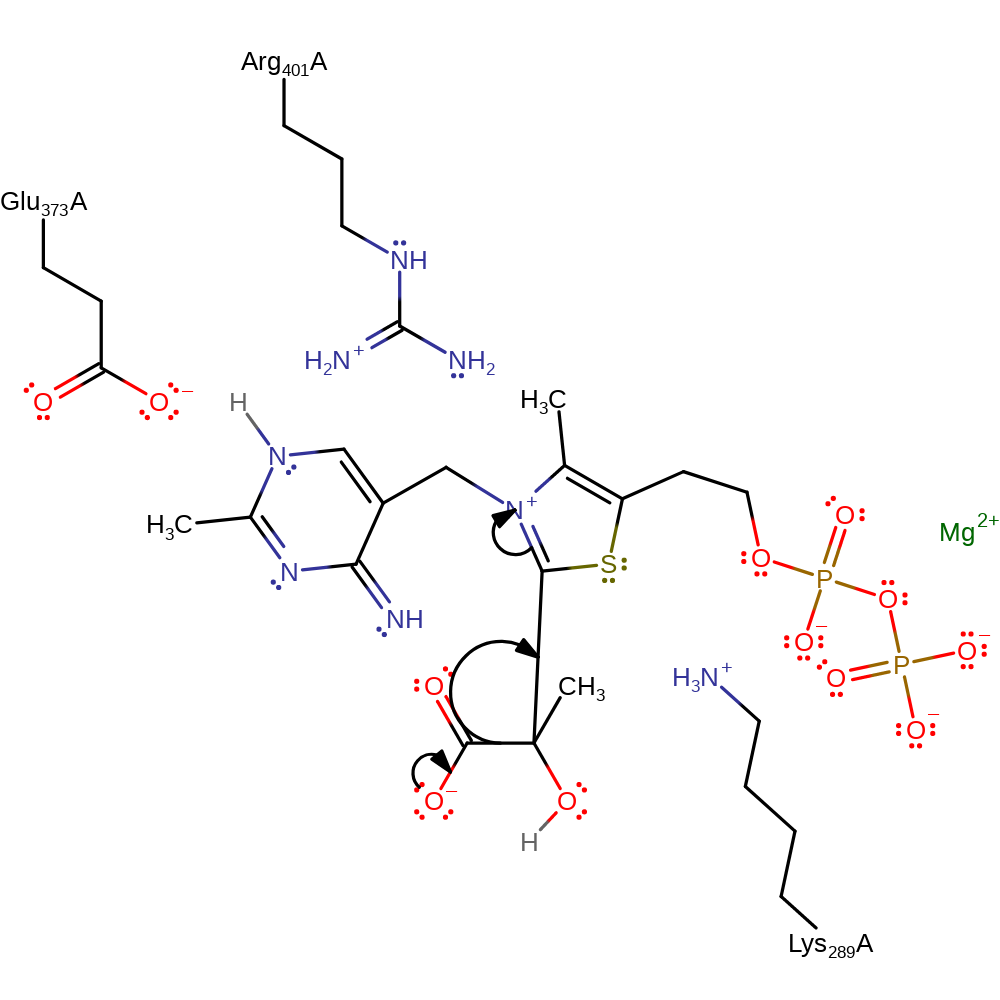
Glu373 deprotonates the N1 position of TDP. Charge delocalisation across the conjugated molecule results in concomitant deprotonation of the thiazole ring, activating the cofactor for the next steps in catalysis. The TDP carbanion, stabilised as an ylid, attacks the carbonyl of pyruvate forming a covalently bound intermediate. The oxyanion intermediate collapses to generate carbon dioxide and a TDP-enol intermediate. The carbanion attacks at the glyceraldehyde group to form a tetrahedral oxyanion intermediate. The tetrahedral intermediate collapses releasing the TDP ylid and 1-deoxy-D-xylulose 5-phosphate, the product which enters the isoprenoid biosynthesis pathway [PMID:19778006]. The cofactor is reprotonated to regenerate the active site.
Catalytic Residues Roles
| UniProt | PDB* (2o1x) | ||
| Glu373 | Glu373A | Activates the thiamine diphosphate cofactor by abstracting a proton, resulting in the production of the active ylid form. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, activator, electrostatic stabiliser |
| Lys289, Arg401 | Lys289A, Arg401A | Act to stabilise the thiamine diphosphate cofactor in its active form. | electrostatic stabiliser |
Chemical Components
proton transfer, cofactor used, bimolecular nucleophilic addition, overall reactant used, intermediate formation, inferred reaction step, intramolecular elimination, overall product formed, intermediate collapse, charge delocalisation, elimination (not covered by the Ingold mechanisms), unimolecular elimination by the conjugate base, native state of cofactor regenerated, native state of enzyme regeneratedReferences
- Xiang S et al. (2007), J Biol Chem, 282, 2676-2682. Crystal Structure of 1-Deoxy-D-xylulose 5-Phosphate Synthase, a Crucial Enzyme for Isoprenoids Biosynthesis. DOI:10.1074/jbc.m610235200. PMID:17135236.
- Frank A et al. (2017), Chem Rev, 117, 5675-5703. The Methylerythritol Phosphate Pathway to Isoprenoids. DOI:10.1021/acs.chemrev.6b00537. PMID:27995802.
- Brammer LA et al. (2009), Org Lett, 11, 4748-4751. Revealing Substrate Promiscuity of 1-Deoxy-d-xylulose 5-Phosphate Synthase. DOI:10.1021/ol901961q. PMID:19778006.
- Shaanan B et al. (2009), FEBS J, 276, 2447-2453. Reaction mechanisms of thiamin diphosphate enzymes: new insights into the role of a conserved glutamate residue. DOI:10.1111/j.1742-4658.2009.06965.x. PMID:19476486.
- Jurgenson CT et al. (2009), Annu Rev Biochem, 78, 569-603. The Structural and Biochemical Foundations of Thiamin Biosynthesis. DOI:10.1146/annurev.biochem.78.072407.102340. PMID:19348578.










 Download:
Download: