Aspartate racemase (CC-type)
Although L-enantiomers of amino acids are predominant in living organisms, D-enantiomers are distributed widely in bacteria, archaea and mammalian tissue. D-aspartate (D-Asp) is one of the most abundant D-enantiomers.
Aspartate racemase (AspR) is a pyridoxal 5'-phosphate (PLP)-independent amino acid racemase that catalyses the interconversion of L-Asp and D-Asp.
Reference Protein and Structure
- Sequence
-
O58403
 (5.1.1.13)
(5.1.1.13)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Pyrococcus horikoshii OT3 (Bacteria)

- PDB
-
1jfl
- CRYSTAL STRUCTURE DETERMINATION OF ASPARTATE RACEMASE FROM AN ARCHAEA
(1.9 Å)



- Catalytic CATH Domains
-
3.40.50.1860
 (see all for 1jfl)
(see all for 1jfl)
Enzyme Mechanism
Introduction
Aspartate racemase (AspR) uses a 'two-base' catalytic mechanism, in which a pair of cysteine residues (Cys82 and Cys194) act as the conjugate acid and base, as seen in other non-PLP dependent alpha-amino acid racemases. The thiolate of Cys82 deprotonates the alpha-C carbon of the amino acid substrate, resulting in the formation of a carbanionic intermediate, which is reprotonated on the reverse side by Cys194, yielding the product with an inverted configuration to the substrate.
Crystallographic studies have identified symmetric, and pseudo-symmetric arrangements of conserved active site residues which are thought to influence the reactivity of aspartate racemase to both D- and L- substrate forms [PMID:12051922]. More recently, crystallographic data which shows the binding of citric acid (a known competitor of aspartate) to the active site has suggested only one of the two cysteine residues is necessary for catalysis [PMID:17847084]. However, the evidence is too inconclusive to ascertain which mechanism is dominant.
Isomerisation continues in both directions until the product dissociates from the active site, therefore, the protonation states of the thiol acid/base pair do not need to be regenerated within the mechanism.
Catalytic Residues Roles
| UniProt | PDB* (1jfl) | ||
| Cys194 | Cys194A | Cys194 acts as the acid, reprotonating the alpha-C of the carbanionic intermediate on the opposite side to the one that the proton was removed from. This results in the formation of the product with the correct inverted configuration. | activator, hydrogen bond donor, proton donor |
| Arg48, Thr84 | Arg48A, Thr84A | Involved in stabilising the negatively charged intermediate. | hydrogen bond donor, electrostatic stabiliser |
| Lys164, Thr124 | Lys164A, Thr124A | Activates and stabilises Cys82. | hydrogen bond donor, electrostatic stabiliser |
| Cys82 | Cys82A | Cys82 (possibly) acts as a base -- some suggestion that it isn't close enough to the substrate -- removing a proton from the alpha-C of the substrate to yield the carbanionic intermediate. | proton acceptor |
Chemical Components
proton transfer, assisted keto-enol tautomerisation, native state of enzyme is not regeneratedReferences
- Ohtaki A et al. (2008), Proteins, 70, 1167-1174. Structure of aspartate racemase complexed with a dual substrate analogue, citric acid, and implications for the reaction mechanism. DOI:10.1002/prot.21528. PMID:17847084.
- Yoshida T et al. (2006), Proteins, 64, 502-512. Roles of conserved basic amino acid residues and activation mechanism of the hyperthermophilic aspartate racemase at high temperature. DOI:10.1002/prot.21010. PMID:16705641.
- Liu L et al. (2002), J Mol Biol, 319, 479-489. Crystal Structure of Aspartate Racemase from Pyrococcus horikoshii OT3 and Its Implications for Molecular Mechanism of PLP-independent Racemization. DOI:10.1016/s0022-2836(02)00296-6. PMID:12051922.
- Yamauchi T et al. (1992), J Biol Chem, 267, 18361-18364. Properties of aspartate racemase, a pyridoxal 5'-phosphate-independent amino acid racemase. PMID:1526977.
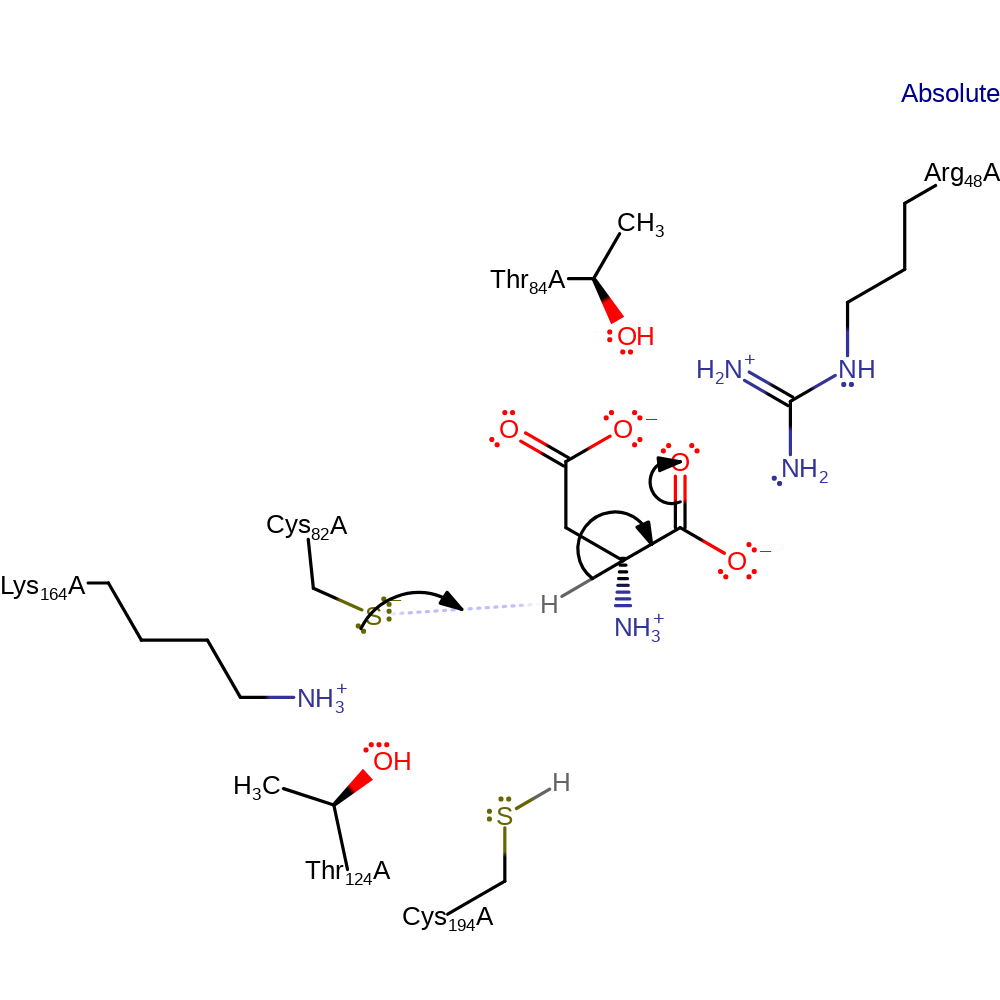
Step 1. Cys82 is thought to exist as a thiolate within the catalytic site, ready to act as a general base upon the C-alpha proton of aspartate. Kinetic studies suggest the mechanism to involve an intermediate, stabilised by the presence of hydrogen bond donating residues and favourable charge-charge interactions.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr124A | hydrogen bond donor, electrostatic stabiliser |
| Arg48A | hydrogen bond donor, electrostatic stabiliser |
| Cys194A | hydrogen bond donor |
| Thr84A | hydrogen bond donor, electrostatic stabiliser |
| Lys164A | electrostatic stabiliser, hydrogen bond donor |
| Cys82A | proton acceptor |
Chemical Components
proton transfer, assisted keto-enol tautomerisation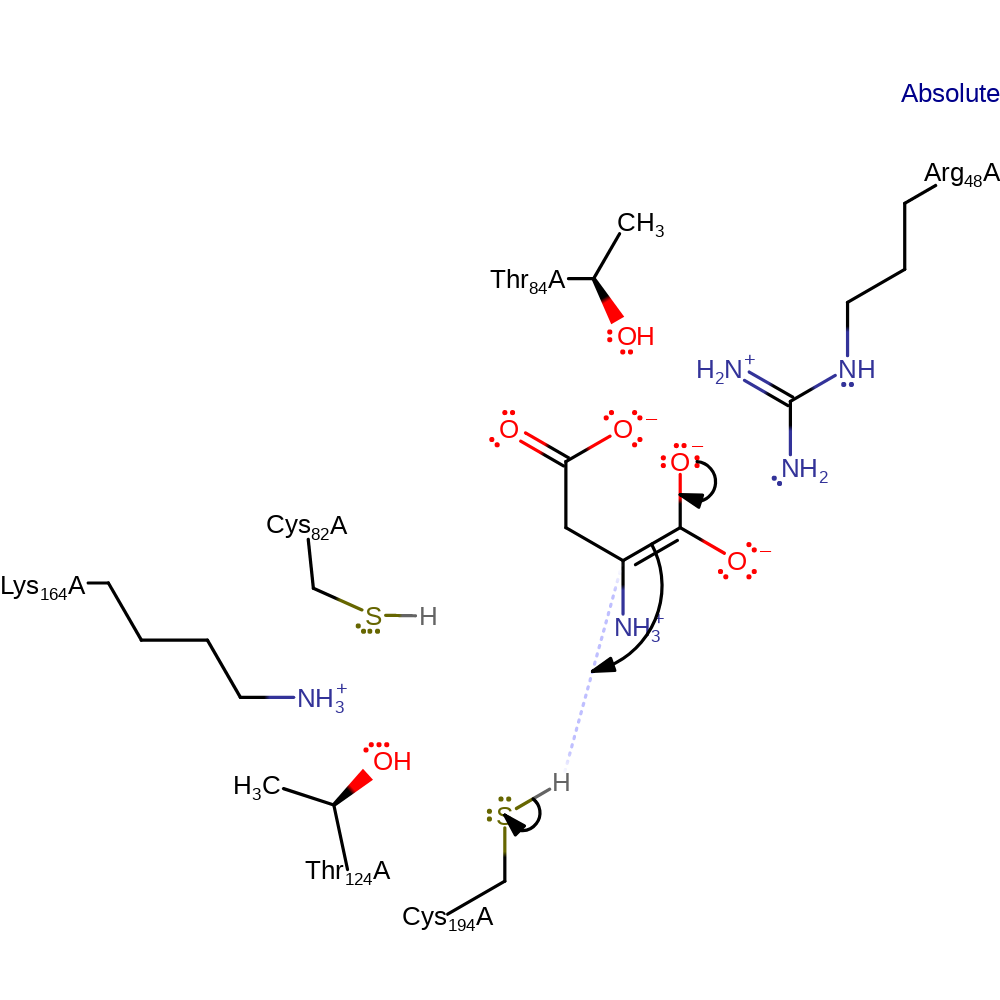
Step 2. The enolate intermediate collapses with concomitant reprotonation from the other face of the molecule. This results in an inversion in stereochemistry.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Thr84A | hydrogen bond donor, electrostatic stabiliser |
| Arg48A | hydrogen bond donor, electrostatic stabiliser |
| Cys194A | activator, hydrogen bond donor |
| Thr124A | hydrogen bond donor, electrostatic stabiliser |
| Lys164A | electrostatic stabiliser, hydrogen bond donor |
| Cys194A | proton donor |

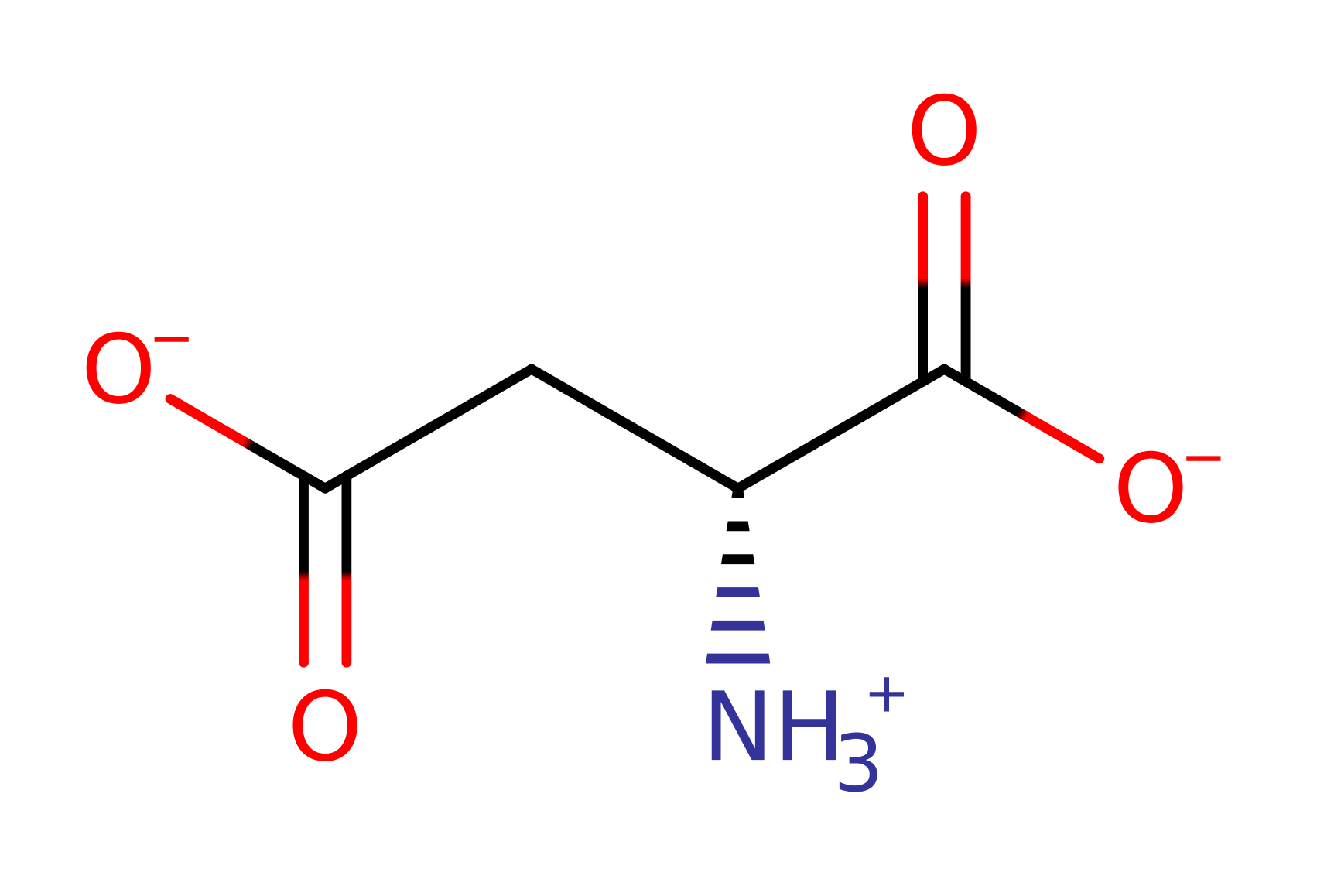
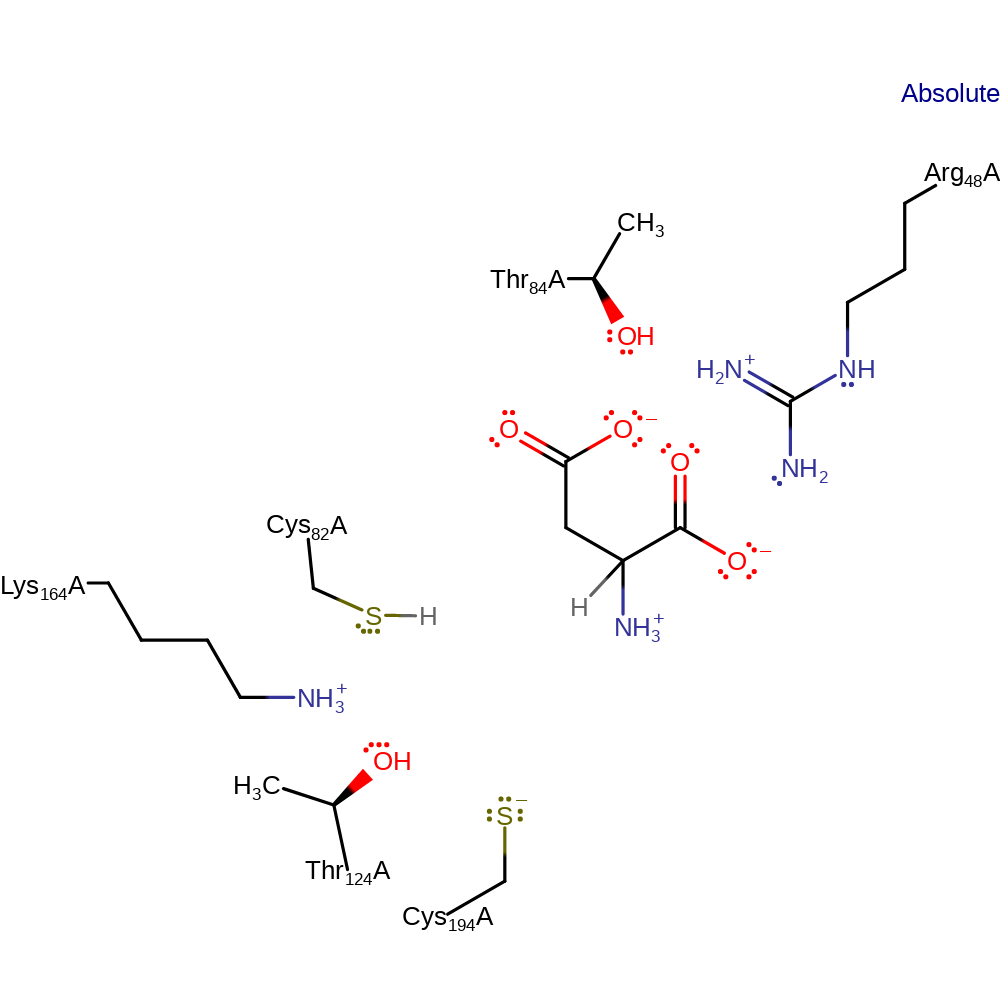 Download:
Download: