Histone-lysine N-methyltransferase (SET7/9 subfamily)
Human methyltransferase SET7/9 catalyses the transfer of a methyl group from the cosubstrate ADOmet to a Lysine residue in Histone H3. Such methylation is used in the regulation of transcription of DNA: it inactivates the DNA towards transcription by creating a silenced chromatin domain. The enzyme displays homology to other SET containing proteins both in humans and in other eukaryotes such as fission yeast (S. cereviseae), with high levels of sequence and structural similarity between the proteins. As a result the SET catalytic domain conserved between the proteins is considered to have the same mechanism in all the methyltransferases.
Several studies have been done on this enzyme, however the method of deprotonation of the histone-lysine is still unclear [PMID:21266482]. Although the mechanism seems to be the same among different proteins, there is a variability in the active site and in presence of zinc finger containing domains. The latter seems to be involved in substrate recognition and binding [PMID:12372304]. Tyr335, Tyr245, and His297 residues are highly conserved [PMID:12372305]. The histidine residues are believed to assist the tyrosine residues with their catalytic function [PMID:12514135]. Tyr305, present only in SET7/9 proteins, appears to assume the role of Tyr335 when this residue is mutated [PMID:20675860]. SET7/9 proteins only perform monomethylation of histone-lysine, whereas other proteins can perform further di- and trimethylations [PMID:12887903]. The rearrangement of water molecules in the active site is proposed to promote this further methylation [8]. The geometry optimisation of SET7/9 complex with SAM substrate showed the formation of CH...O hydrogen bonds AdoMet methyl and His293 main chain carbonyl and Tyr335 hydroxyl [PMID:21454678]. The unusually high pH optimum [PMID:12887903, PMID:12372305] suggests partial deprotonation of the target lysine and hydroxyl group of the tyrosine. Nonetheless, pKa calculations [PMID:18391193] contradict the presence of tyrosine hydroxyl anion.
Reference Protein and Structure
- Sequence
-
Q8WTS6
 (2.1.1.364)
(2.1.1.364)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Homo sapiens (Human)

- PDB
-
1h3i
- Crystal structure of the Histone Methyltransferase SET7/9
(2.1 Å)



- Catalytic CATH Domains
-
2.170.270.10
 (see all for 1h3i)
(see all for 1h3i)
Enzyme Reaction (EC:2.1.1.43)
Enzyme Mechanism
Introduction
Proceeds via in-line nucleophilic attack from the target lysine residue in the substrate on the methyl group of the ADOmet molecule, with the lysine activated to act as a nucleophile by deprotonation from a Tyr 335-His 297 diad. This transfers the methyl group to the lysine.
Catalytic Residues Roles
| UniProt | PDB* (1h3i) | ||
| His297 | His297(246)A | Forms a dyad with Tyr335, activating this residue to act as a general acid/base. | hydrogen bond acceptor, electrostatic stabiliser |
| Tyr335 | Tyr335(284)A | Acts as general base to deprotonate the lysine residue in the target histone such that it can act as a nucleophile. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, activator, electrostatic stabiliser |
| His293 | His293(242)A | Acts to ensure that Tyr 335 is in the correct protonation state by participating in a proton relay system whereby the proton from the substrate lysine is transferred via Tyr 335 to the residue. | hydrogen bond acceptor, electrostatic stabiliser |
| Tyr305, Tyr245 | Tyr305(254)A, Tyr245(194)A | Help stabilise the reactive intermediates and transition states formed during the course of the reaction. | activator, hydrogen bond acceptor, electrostatic stabiliser |
Chemical Components
proton transfer, intermediate formation, overall reactant used, overall product formed, bimolecular nucleophilic substitution, intermediate terminated, cofactor used, native state of enzyme regeneratedReferences
- Kwon T et al. (2003), EMBO J, 22, 292-303. Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet. DOI:10.1093/emboj/cdg025. PMID:12514135.
- Luo M (2018), Chem Rev, 118, 6656-6705. Chemical and Biochemical Perspectives of Protein Lysine Methylation. DOI:10.1021/acs.chemrev.8b00008. PMID:29927582.
- Cortopassi WA et al. (2016), J Mol Graph Model, 67, 69-84. Mechanisms of histone lysine-modifying enzymes: A computational perspective on the role of the protein environment. DOI:10.1016/j.jmgm.2016.04.011. PMID:27258188.
- Horowitz S et al. (2011), J Biol Chem, 286, 18658-18663. Direct Evidence for Methyl Group Coordination by Carbon-Oxygen Hydrogen Bonds in the Lysine Methyltransferase SET7/9. DOI:10.1074/jbc.m111.232876. PMID:21454678.
- Xu S et al. (2011), Nucleic Acids Res, 39, 4438-4449. Structural and biochemical studies of human lysine methyltransferase Smyd3 reveal the important functional roles of its post-SET and TPR domains and the regulation of its activity by DNA binding. DOI:10.1093/nar/gkr019. PMID:21266482.
- Del Rizzo PA et al. (2010), J Biol Chem, 285, 31849-31858. SET7/9 Catalytic Mutants Reveal the Role of Active Site Water Molecules in Lysine Multiple Methylation. DOI:10.1074/jbc.m110.114587. PMID:20675860.
- Zhang X et al. (2008), Proc Natl Acad Sci U S A, 105, 5728-5732. Enzymatic mechanism and product specificity of SET-domain protein lysine methyltransferases. DOI:10.1073/pnas.0801788105. PMID:18391193.
- Hu P et al. (2006), J Am Chem Soc, 128, 1272-1278. Catalytic mechanism and product specificity of the histone lysine methyltransferase SET7/9: an ab initio QM/MM-FE study with multiple initial structures. DOI:10.1021/ja056153+. PMID:16433545.
- Zhang X et al. (2003), Mol Cell, 12, 177-185. Structural basis for the product specificity of histone lysine methyltransferases. DOI:10.2210/pdb1peg/pdb. PMID:12887903.
- Trievel RC et al. (2002), Cell, 111, 91-103. Structure and catalytic mechanism of a SET domain protein methyltransferase. DOI:10.2210/pdb1mlv/pdb. PMID:12372303.
- Zhang X et al. (2002), Cell, 111, 117-127. Structure of the Neurospora SET domain protein DIM-5, a histone H3 lysine methyltransferase. DOI:10.2210/pdb1ml9/pdb. PMID:12372305.
- Wilson JR et al. (2002), Cell, 111, 105-115. Crystal Structure and Functional Analysis of the Histone Methyltransferase SET7/9. DOI:10.1016/s0092-8674(02)00964-9. PMID:12372304.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| His293(242)A | electrostatic stabiliser, hydrogen bond acceptor |
| Tyr245(194)A | activator, electrostatic stabiliser, hydrogen bond acceptor, hydrogen bond donor |
| Tyr335(284)A | activator, electrostatic stabiliser, hydrogen bond acceptor, hydrogen bond donor |
| Tyr305(254)A | activator, electrostatic stabiliser, hydrogen bond acceptor |
| His297(246)A | electrostatic stabiliser, hydrogen bond acceptor |
| Tyr335(284)A | proton acceptor |
Chemical Components
proton transfer, intermediate formation, overall reactant used, overall product formed
Step 2. The histone-lysine attacks the SAM substrate in a nucleophilic substitution reaction, resulting in the transfer of the methyl group from SAM to the histone.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His293(242)A | electrostatic stabiliser, hydrogen bond acceptor |
| Tyr245(194)A | activator, electrostatic stabiliser, hydrogen bond acceptor, hydrogen bond donor |
| Tyr335(284)A | hydrogen bond acceptor, electrostatic stabiliser, activator, hydrogen bond donor |
| Tyr305(254)A | activator, electrostatic stabiliser, hydrogen bond acceptor |
| His297(246)A | electrostatic stabiliser, hydrogen bond acceptor |
| Tyr335(284)A | proton donor |



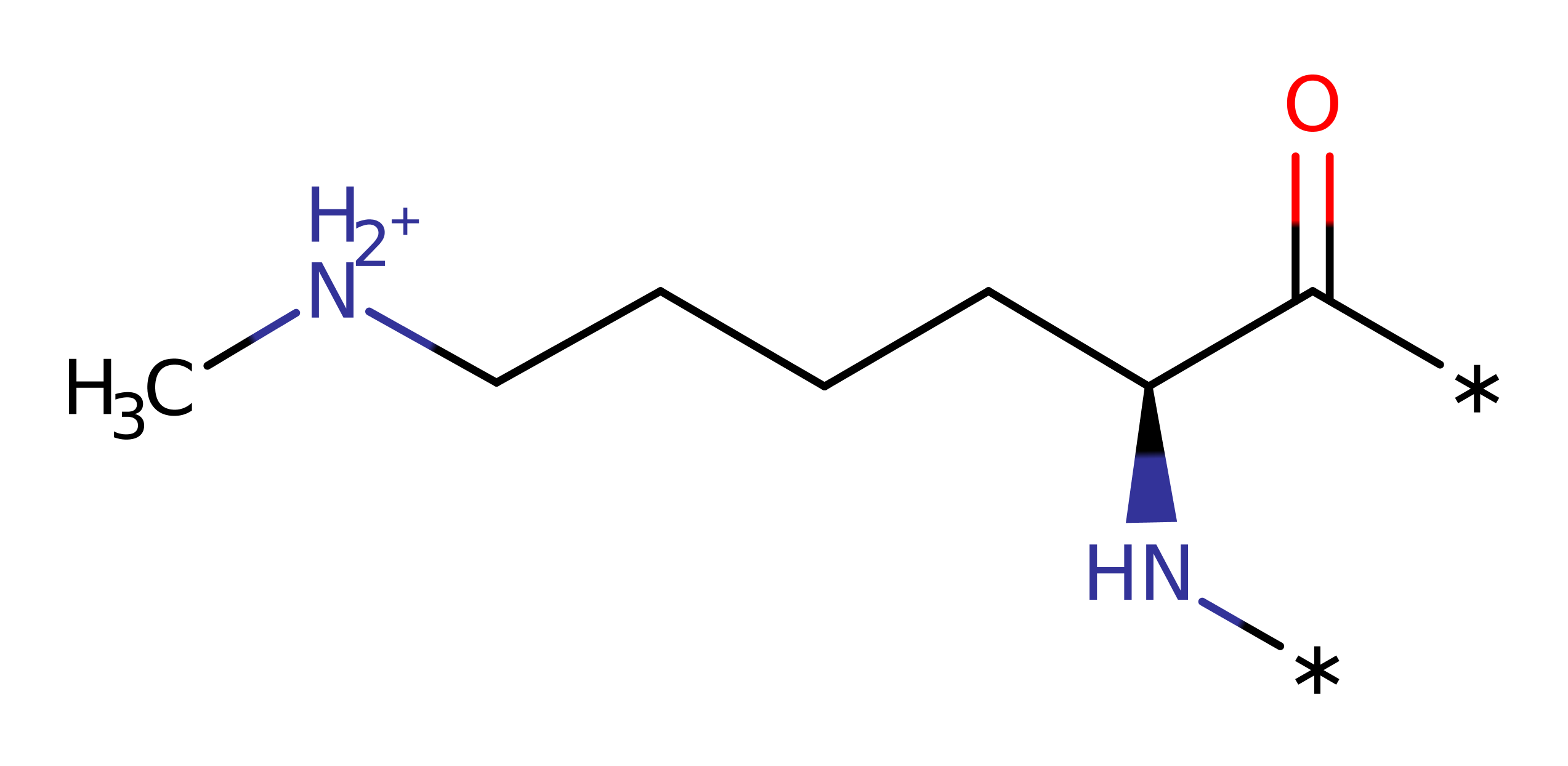


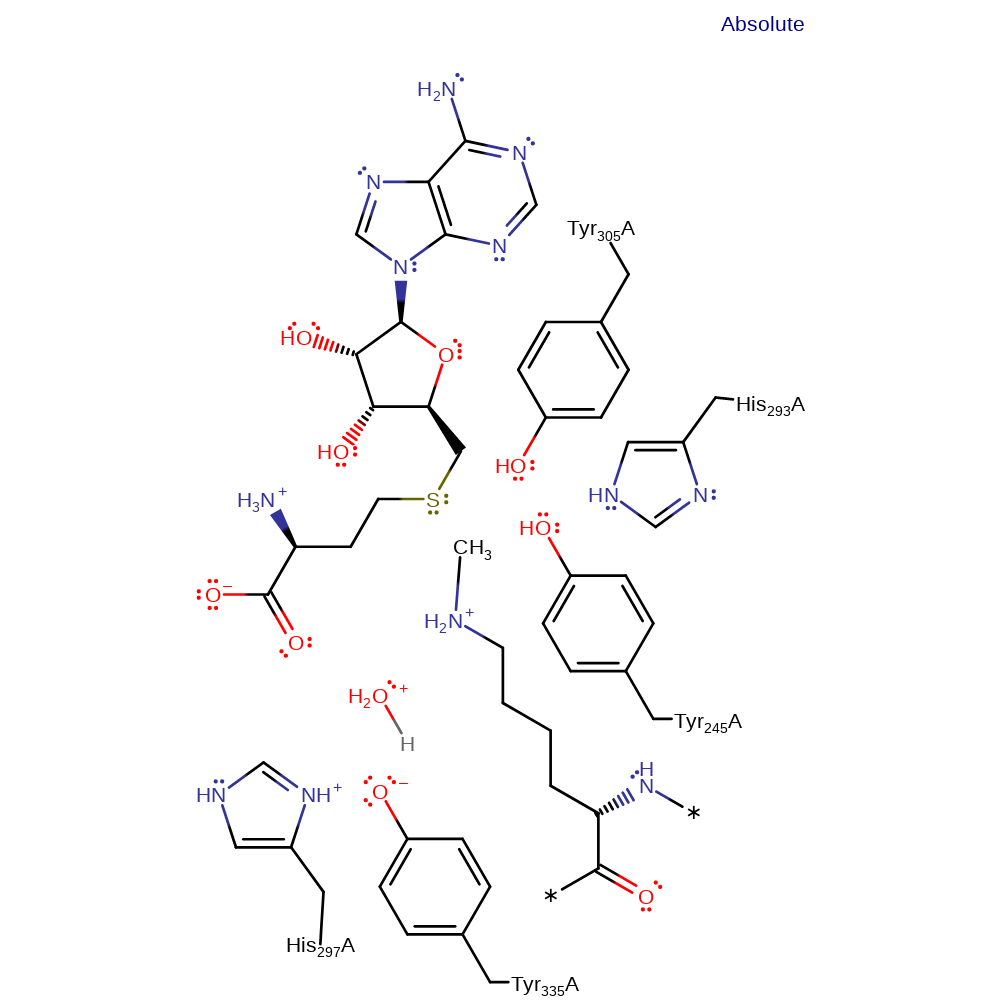 Download:
Download: