(S)-2-haloacid dehalogenase
(S)-2-haloacid dehalogenase catalyses the hydrolytic dehalogenation of L-2-haloacid to the corresponding D-2-hydroxyacid with the inversion of the configuration at the C2 atom. The degradation of halogenated hydrocarbons used in large quantities to make solvents and plastics. The stereospecificity of this enzyme is of interest for its potential use in the biosynthesis of chiral compounds. It is a member of the haloacid dehalogenase superfamily.
Reference Protein and Structure
- Sequence
-
Q60099
 (3.8.1.2)
(3.8.1.2)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Xanthobacter autotrophicus (Bacteria)

- PDB
-
1qq5
- STRUCTURE OF L-2-HALOACID DEHALOGENASE FROM XANTHOBACTER AUTOTROPHICUS
(1.52 Å)



- Catalytic CATH Domains
-
3.40.50.1000
 1.10.150.240
1.10.150.240  (see all for 1qq5)
(see all for 1qq5)
- Cofactors
- Water (1)
Enzyme Reaction (EC:3.8.1.2)
Enzyme Mechanism
Introduction
When the L-2-haloacid is taken into the active site its carboxyl group is recognised by Arg41 before forming a Michaelis compound with the enzyme. The substrate binding drives out a water molecule freeing Asp10's nucleophilic oxygen allowing attack on the C2 carbon via an SN2 mechanism. Asp10 is stabilised by hydrogen bonding to Lys151, Thr14 and Ser175. Arg41 is thought to abstract the halide ion from the substrate. Ser118 serves as the main residue for stabilising the substrate carboxyl moiety in the reaction intermediate. Asn177 and Asp180 are also thought to be involved in the hydrolysis reaction.
Catalytic Residues Roles
| UniProt | PDB* (1qq5) | ||
| Asp8 | Asp8A | Acts as the catalytic nucleophile, forming a covalently bound intermediate with the substrate. | hydrogen bond acceptor, nucleofuge, nucleophile |
| Asp176 | Asp176A | Acts as a general acid/base. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor, activator |
| Arg39, Phe175, Asn115 | Arg39A, Phe175A, Asn115A | Stabilise the chloride leaving group. | hydrogen bond donor, electrostatic stabiliser |
| Lys147, Thr12, Ser171, Asn173, Ser114 | Lys147A, Thr12A, Ser171A, Asn173A, Ser114A | Activate the substrate. | activator, hydrogen bond donor |
Chemical Components
bimolecular nucleophilic substitution, atom stereo change, overall reactant used, overall product formed, enzyme-substrate complex formation, intermediate formation, proton transfer, enzyme-substrate complex cleavage, intermediate terminated, intermediate collapse, hydrolysis, native state of enzyme regenerated, inferred reaction stepReferences
- Li YF et al. (1998), J Biol Chem, 273, 15035-15044. Crystal Structures of Reaction Intermediates ofL-2-Haloacid Dehalogenase and Implications for the Reaction Mechanism. DOI:10.1074/jbc.273.24.15035. PMID:9614112.
- Kumar A et al. (2016), Int J Biol Macromol, 83, 216-225. l-2-Haloacid dehalogenase from Ancylobacter aquaticus UV5: Sequence determination and structure prediction. DOI:10.1016/j.ijbiomac.2015.11.066. PMID:26645146.
- Paneth P (2003), Acc Chem Res, 36, 120-126. Chlorine Kinetic Isotope Effects on Enzymatic Dehalogenations. DOI:10.1021/ar010101h. PMID:12589697.
- Lahiri SD et al. (2002), Biochemistry, 41, 8351-8359. Caught in the Act: The Structure of Phosphorylatedβ-Phosphoglucomutase fromLactococcus lactis†,‡. DOI:10.1021/bi0202373.
- Ichiyama S et al. (2000), J Biol Chem, 275, 40804-40809. Novel Catalytic Mechanism of Nucleophilic Substitution by Asparagine Residue Involving Cyanoalanine Intermediate Revealed by Mass Spectrometric Monitoring of an Enzyme Reaction. DOI:10.1074/jbc.m008065200. PMID:11006296.
- Morais MC et al. (2000), Biochemistry, 39, 10385-10396. The Crystal Structure ofBacillus cereusPhosphonoacetaldehyde Hydrolase: Insight into Catalysis of Phosphorus Bond Cleavage and Catalytic Diversification within the HAD Enzyme Superfamily†,‡. DOI:10.1021/bi001171j.
- Ridder IS et al. (1999), J Biol Chem, 274, 30672-30678. Crystal Structures of Intermediates in the Dehalogenation of Haloalkanoates by L-2-Haloacid Dehalogenase. DOI:10.1074/jbc.274.43.30672. PMID:10521454.
- Kurihara T et al. (1995), J Biochem, 117, 1317-1322. Comprehensive site-directed mutagenesis of L-2-halo acid dehalogenase to probe catalytic amino acid residues. PMID:7490277.
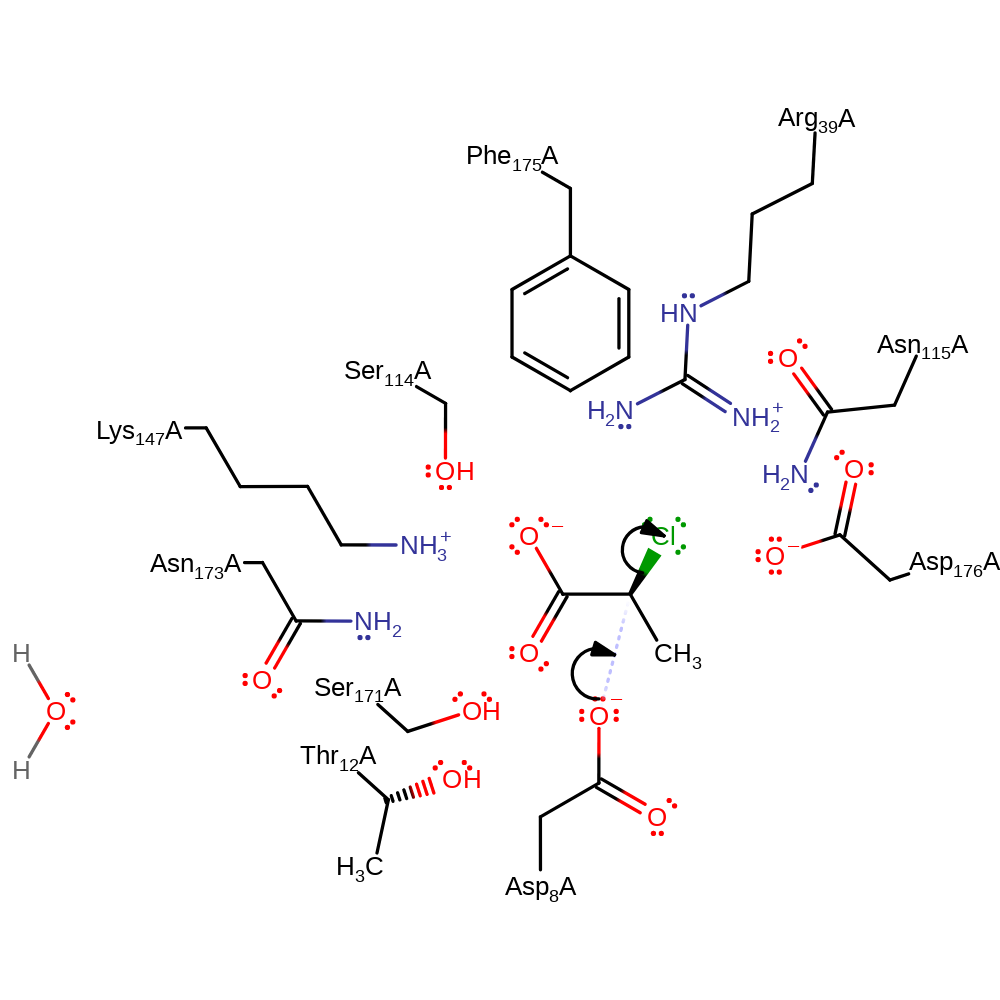
Step 1. Asp8 initiates a nucleophilic attack upon the chlorinated carbon of the substrate in a substitution reaction, eliminating chloride.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp176A | hydrogen bond acceptor, activator |
| Arg39A | hydrogen bond donor |
| Lys147A | hydrogen bond donor, activator |
| Ser171A | hydrogen bond donor, electrostatic stabiliser |
| Thr12A | hydrogen bond donor, electrostatic stabiliser |
| Asn173A | hydrogen bond donor, electrostatic stabiliser |
| Asp8A | hydrogen bond acceptor |
| Phe175A | polar/non-polar interaction |
| Asn115A | hydrogen bond donor |
| Ser114A | electrostatic stabiliser |
| Asp8A | nucleophile |
Chemical Components
ingold: bimolecular nucleophilic substitution, atom stereo change, overall reactant used, overall product formed, enzyme-substrate complex formation, intermediate formation
Step 2. Asp176 deprotonates water, which initiates a nucleophilic attack on the carbonyl of Asp8, as demonstrated by isotope studies, eliminating Asp8 while maintaining R stereochemistry, which is inverted to that of the S-haloacid substrate.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp176A | hydrogen bond acceptor |
| Arg39A | hydrogen bond donor, electrostatic stabiliser |
| Lys147A | hydrogen bond donor |
| Ser171A | hydrogen bond donor, electrostatic stabiliser |
| Asn115A | hydrogen bond donor, hydrogen bond acceptor, electrostatic stabiliser |
| Thr12A | hydrogen bond donor, electrostatic stabiliser |
| Asn173A | hydrogen bond donor, electrostatic stabiliser |
| Asp8A | hydrogen bond acceptor |
| Phe175A | polar/non-polar interaction, electrostatic stabiliser |
| Ser114A | electrostatic stabiliser |
| Asp8A | nucleofuge |
| Asp176A | proton acceptor |
Chemical Components
proton transfer, ingold: bimolecular nucleophilic substitution, overall product formed, enzyme-substrate complex cleavage, intermediate terminated, intermediate collapse, hydrolysisCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp176A | hydrogen bond donor |
| Lys147A | hydrogen bond donor |
| Ser171A | hydrogen bond donor, electrostatic stabiliser |
| Thr12A | hydrogen bond donor, electrostatic stabiliser |
| Asn173A | hydrogen bond donor, electrostatic stabiliser |
| Asp8A | hydrogen bond acceptor |
| Asp176A | proton donor |






 Download:
Download: