Nitrite reductase (copper type)
This family of enzymes is found in both denitrifying bacteria and fungi which reside in soil and aquatic ecosystems.
Denitrification, the reduction of dissolved nitrate and nitrite to gaseous NO, N20 and N2 is an important process in the recycling of nitrogen in the biosphere, and is a key step in the nitrogen cycle resulting in the loss of terrestrial nitrogen to the atmosphere.
It is chiefly carried out by denitrifying bacteria, which contain nitrate and nitrite reductases. The copper-containing family of enzymes is found in both denitrifying bacteria and fungi which reside in soil and aquatic ecosystems and uses copper ions as cofactors rather than the more common haem cd1
The enzymes in question catalyse the reduction of nitrite (NO2-) to NO + H2O. They contain two copper centres, a Type I centre which receives electrons from pseudoazurin (a copper containing protein), and a type II centre which is the site of nitrite reduction.
Reference Protein and Structure
- Sequence
-
P25006
 (1.7.2.1)
(1.7.2.1)
 (Sequence Homologues)
(PDB Homologues)
(Sequence Homologues)
(PDB Homologues)
- Biological species
-
Achromobacter cycloclastes (Bacteria)

- PDB
-
1nia
- THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED
(2.5 Å)



- Catalytic CATH Domains
-
2.60.40.420
 (see all for 1nia)
(see all for 1nia)
- Cofactors
- Copper(2+) (2) Metal MACiE
Enzyme Reaction (EC:1.7.2.1)
Enzyme Mechanism
Introduction
The resting oxidised state of the enzyme contains a water molecule coordinated to a Cu2+ ion at the type II copper centre. Nitrite then binds to the type II copper ion and and displaces the water molecule. This makes electron transfer from pseudoazurin to the type II copper centre via the type I copper centre energetically favourable. Nitrite reduction occurs at the type II centre. The type II copper transfers the electron it received to the nitrite, which together with protonation of the nitrite leads to reduction of nitrite to NO + H2O.
Two protons are needed for the reduction of NO2- to NO + H2O, which are likely to come from bulk solvent as the Asp98A catalytic residue is at the end of a proton tunnel connected to the bulk solvent.
Catalytic Residues Roles
| UniProt | PDB* (1nia) | ||
| Asp136 | Asp98A | Involved in supplying proton(s) for the reduction of NO2- to NO + H2O. | activator, hydrogen bond acceptor, proton acceptor, proton donor |
| His173 | His135A | Binds the Cu(II) ion in the type II site and is part of the electron transfer pathway to the Cu(II) type II site from the type I site. | single electron relay, single electron acceptor, single electron donor, metal ligand |
| Cys174 | Cys136A | Binds the Cu(II) ion in the type I site and is part of the electron transfer pathway to the Cu(II) type II site from the type I site. | single electron relay, single electron acceptor, single electron donor, metal ligand |
| His293 | His255B | Has been variously proposed to have roles in providing a positive charge to assist the pushing of an electron onto the nitrite during the reduction, protonating the substrate during the reduction, and modifying the properties of Asp 98 via the intervening water molecule. | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Glu317 (main-C), Thr318 | Glu279B (main-C), Thr280B | Activates His255. | electrostatic stabiliser |
| His183, Met188, His133 | His145A, Met150A, His95A | Binds the Cu(II) ion in the type I site. | metal ligand |
| His344, His138 | His306B, His100A | Binds the Cu(II) ion in the type II site. | metal ligand |
Chemical Components
overall reactant used, intermediate formation, proton transfer, electron transferReferences
- Fukuda Y et al. (2016), Proc Natl Acad Sci U S A, 113, 2928-2933. Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography. DOI:10.1073/pnas.1517770113. PMID:26929369.
- Li Y et al. (2015), Biochemistry, 54, 1233-1242. Enzymatic Mechanism of Copper-Containing Nitrite Reductase. DOI:10.1021/bi5007767. PMID:25594136.
- Impagliazzo A et al. (2005), Chembiochem, 6, 1648-1653. Pseudoazurin-Nitrite Reductase Interactions. DOI:10.1002/cbic.200500082. PMID:16138306.
- Astier Y et al. (2005), Chemphyschem, 6, 1114-1120. Sensing Nitrite through a Pseudoazurin-Nitrite Reductase Electron Transfer Relay. DOI:10.1002/cphc.200400384. PMID:15900523.
- Kataoka K et al. (2004), J Biol Chem, 279, 53374-53378. Structure-based Engineering of Alcaligenes xylosoxidans Copper-containing Nitrite Reductase Enhances Intermolecular Electron Transfer Reaction with Pseudoazurin. DOI:10.1074/jbc.m410198200. PMID:15475344.
- Pinho D et al. (2004), Eur J Biochem, 271, 2361-2369. Copper-containing nitrite reductase from Pseudomonas chlororaphis DSM 50135. Evidence for modulation of the rate of intramolecular electron transfer through nitrite binding to the type 2 copper center. DOI:10.1111/j.1432-1033.2004.04155.x. PMID:15182351.
- Boulanger MJ et al. (2000), J Biol Chem, 275, 23957-23964. Catalytic Roles for Two Water Bridged Residues (Asp-98 and His-255) in the Active Site of Copper-containing Nitrite Reductase. DOI:10.1074/jbc.m001859200. PMID:10811642.
- Suzuki S et al. (2000), Acc Chem Res, 33, 728-735. Metal Coordination and Mechanism of Multicopper Nitrite Reductase. DOI:10.1021/ar9900257. PMID:11041837.
- Murphy ME et al. (1997), J Biol Chem, 272, 28455-28460. Structure of Nitrite Bound to Copper-containing Nitrite Reductase from Alcaligenes faecalis: MECHANISTIC IMPLICATIONS. DOI:10.1074/jbc.272.45.28455. PMID:9353305.
- Adman ET et al. (1995), J Biol Chem, 270, 27458-27474. The Structure of Copper-nitrite Reductase from Achromobacter cycloclastes at Five pH Values, with NO(2)[IMAGE] Bound and with Type II Copper Depleted. DOI:10.1074/jbc.270.46.27458. PMID:7499203.
- Kukimoto M et al. (1994), Biochemistry, 33, 5246-5252. X-ray Structure and Site-Directed Mutagenesis of a Nitrite Reductase from Alcaligenes Faecalis S-6: Roles of Two Copper Atoms in Nitrite Reduction. DOI:10.1021/bi00183a030. PMID:8172899.
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp98A | hydrogen bond acceptor, activator |
| His135A | metal ligand |
| Cys136A | metal ligand |
| His255B | hydrogen bond donor |
| His306B | metal ligand |
| His100A | metal ligand |
| His145A | metal ligand |
| Met150A | metal ligand |
| His95A | metal ligand |
| Asp98A | proton acceptor |
Chemical Components
overall reactant used, intermediate formation, proton transferCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp98A | hydrogen bond acceptor |
| His135A | metal ligand |
| Cys136A | metal ligand |
| His255B | hydrogen bond donor |
| His145A | metal ligand |
| His95A | metal ligand |
| Met150A | metal ligand |
| His100A | metal ligand |
| His306B | metal ligand |
| Asp98A | proton donor |
Chemical Components
proton transfer
Step 3. A single electron is transferred from Pseudoazurin, via the type I copper ion, Cys136, and His135 to the type II copper ion.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp98A | hydrogen bond acceptor |
| His135A | metal ligand |
| Cys136A | metal ligand |
| His255B | hydrogen bond acceptor |
| His100A | metal ligand |
| His306B | metal ligand |
| His145A | metal ligand |
| His95A | metal ligand |
| Met150A | metal ligand |
| Cys136A | single electron donor |
| His135A | single electron relay |
| Cys136A | single electron relay |
| His135A | single electron donor |
| Cys136A | single electron acceptor |
| His135A | single electron acceptor |
Chemical Components
electron transferCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His306B | metal ligand |
| His100A | metal ligand |
| His145A | metal ligand |
| His95A | metal ligand |
| Met150A | metal ligand |
| His135A | metal ligand |
| Cys136A | metal ligand |
Chemical Components
Catalytic Residues Roles
| Residue | Roles |
|---|---|
| His145A | metal ligand |
| His95A | metal ligand |
| Met150A | metal ligand |
| His306B | metal ligand |
| His100A | metal ligand |
| Cys136A | metal ligand |
| His135A | metal ligand |
| His255B | proton donor |
Chemical Components
proton transfer
Step 6. The nitrous acid substrate collapses to copper bound N=O and water by deprotonating the activated water molecule.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His100A | metal ligand |
| His306B | metal ligand |
| His145A | metal ligand |
| His95A | metal ligand |
| Met150A | metal ligand |
| His135A | metal ligand |
| Cys136A | metal ligand |
| Thr280B | electrostatic stabiliser |
| Glu279B (main-C) | electrostatic stabiliser |
Chemical Components
proton transfer
Step 7. The type II copper ion donates an electron to the NO+ intermediate, forming the final product (NO radical).
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His145A | metal ligand |
| His95A | metal ligand |
| Met150A | metal ligand |
| His100A | metal ligand |
| His306B | metal ligand |
| His135A | metal ligand |
| Cys136A | metal ligand |
| Thr280B | electrostatic stabiliser |
| Glu279B (main-C) | electrostatic stabiliser |
Chemical Components
electron transfer
Step 8. Water displaces the NO product and His255 rotates back to its starting position with reprotonation.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| His306B | metal ligand |
| His100A | metal ligand |
| Met150A | metal ligand |
| His95A | metal ligand |
| His145A | metal ligand |
| His135A | metal ligand |
| Cys136A | metal ligand |
| His255B | proton acceptor |
Chemical Components
proton transferIntroduction
The resting oxidised state of the enzyme contains a water molecule coordinated to a Cu2+ ion at the type II copper centre. Nitrite then binds to the type II copper ion and and displaces the water molecule. The type II copper now receives an electron from the type I copper and transfers it to the nitrite, which together with protonation of the nitrite leads to reduction of nitrite to NO + H2O.
Two protons are needed for the reduction of NO2- to NO + H2O, but the details of proton transfer have still not been firmly established. Proposed sources for the protons include His 255 (although it has also been proposed that this residue is not positioned appropriately for this role); the water molecule that bridges His 255 and Asp 98; the water molecule initially bound to copper (which is suggested to be deprotonated by Asp 98 prior to its displacement by NO2-, with Asp 98 later supplying the proton to the nitrite); and the nitrite entering as HNO2 with one proton already present.
Catalytic Residues Roles
| UniProt | PDB* (1nia) | ||
| Asp136 | Asp98A | Activates His255 to act as the general acid/base through a water molecule. | activator, hydrogen bond acceptor |
| His173 | His135A | Binds the Cu(II) ion in the type II site and is part of the electron transfer pathway from the type I Cu(II) site to the type II Cu(II) site. | single electron relay, single electron acceptor, single electron donor, metal ligand |
| Cys174 | Cys136A | Binds the Cu(II) ion in the type I site and is part of the electron transfer pathway from the type I Cu(II) site to the type II Cu(II) site. | single electron relay, single electron acceptor, single electron donor, metal ligand |
| His293 | His255B | General acid/base | hydrogen bond acceptor, hydrogen bond donor, proton acceptor, proton donor |
| Glu317 (main-C), Thr318 | Glu279B (main-C), Thr280B | Help modify the pKa of His255. | modifies pKa |
| His183, Met188, His133 | His145A, Met150A, His95A | Bind the Cu(II) ion in the type I site. | metal ligand |
| His344, His138 | His306B, His100A | Bind the Cu(II) ion in the type II site. | metal ligand |
Chemical Components
electron transfer, coordination, overall reactant used, coordination to a metal ion, intermediate formation, electron relay, radical formation, redox reaction, elimination (not covered by the Ingold mechanisms), proton transfer, heterolysis, overall product formed, dehydration, decoordination from a metal ion, native state of enzyme regeneratedReferences
- Suzuki S et al. (2000), Acc Chem Res, 33, 728-735. Metal Coordination and Mechanism of Multicopper Nitrite Reductase. DOI:10.1021/ar9900257. PMID:11041837.
- Antonyuk SV et al. (2005), Proc Natl Acad Sci U S A, 102, 12041-12046. Atomic resolution structures of resting-state, substrate- and product-complexed Cu-nitrite reductase provide insight into catalytic mechanism. DOI:10.1073/pnas.0504207102. PMID:16093314.
- Tocheva EI et al. (2004), Science, 304, 867-870. Side-On Copper-Nitrosyl Coordination by Nitrite Reductase. DOI:10.1126/science.1095109. PMID:15131305.
- Boulanger MJ et al. (2000), J Biol Chem, 275, 23957-23964. Catalytic Roles for Two Water Bridged Residues (Asp-98 and His-255) in the Active Site of Copper-containing Nitrite Reductase. DOI:10.1074/jbc.m001859200. PMID:10811642.
- Inoue T et al. (1999), J Biol Chem, 274, 17845-17852. Crystal Structure Determinations of Oxidized and Reduced Pseudoazurins from Achromobacter cycloclastes: CONCERTED MOVEMENT OF COPPER SITE IN REDOX FORMS WITH THE REARRANGEMENT OF HYDROGEN BOND AT A REMOTE HISTIDINE. DOI:10.1074/jbc.274.25.17845. PMID:10364229.
- Adman ET et al. (1995), J Biol Chem, 270, 27458-27474. The Structure of Copper-nitrite Reductase from Achromobacter cycloclastes at Five pH Values, with NO(2)[IMAGE] Bound and with Type II Copper Depleted. DOI:10.1074/jbc.270.46.27458. PMID:7499203.
- Kukimoto M et al. (1994), Biochemistry, 33, 5246-5252. X-ray Structure and Site-Directed Mutagenesis of a Nitrite Reductase from Alcaligenes Faecalis S-6: Roles of Two Copper Atoms in Nitrite Reduction. DOI:10.1021/bi00183a030. PMID:8172899.

Step 1. Pseudoazurin donates an electron, through the type I copper ion, Cys136 and His135 to the type II copper ion.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp98A | hydrogen bond acceptor, activator |
| His135A | metal ligand |
| Cys136A | metal ligand |
| His255B | hydrogen bond donor |
| Cys136A | single electron acceptor |
| His100A | metal ligand |
| His306B | metal ligand |
| His145A | metal ligand |
| His95A | metal ligand |
| Met150A | metal ligand |
| His135A | single electron acceptor |
| Cys136A | single electron donor, single electron relay |
| His135A | single electron relay, single electron donor |
Chemical Components
electron transfer, coordination, overall reactant used, coordination to a metal ion, intermediate formation, electron relay
Step 2. Cu502 donates the electron to the bound nitrous acid, which collapses to form water, deprotonating His255B, and a Cu-bound NO radical.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp98A | hydrogen bond acceptor |
| His135A | metal ligand |
| Cys136A | metal ligand |
| His255B | hydrogen bond donor |
| Glu279B (main-C) | modifies pKa |
| Thr280B | modifies pKa |
| His255B | proton donor |
Chemical Components
radical formation, redox reaction, elimination (not covered by the Ingold mechanisms), proton transfer, heterolysis, overall product formed, dehydration, decoordination from a metal ion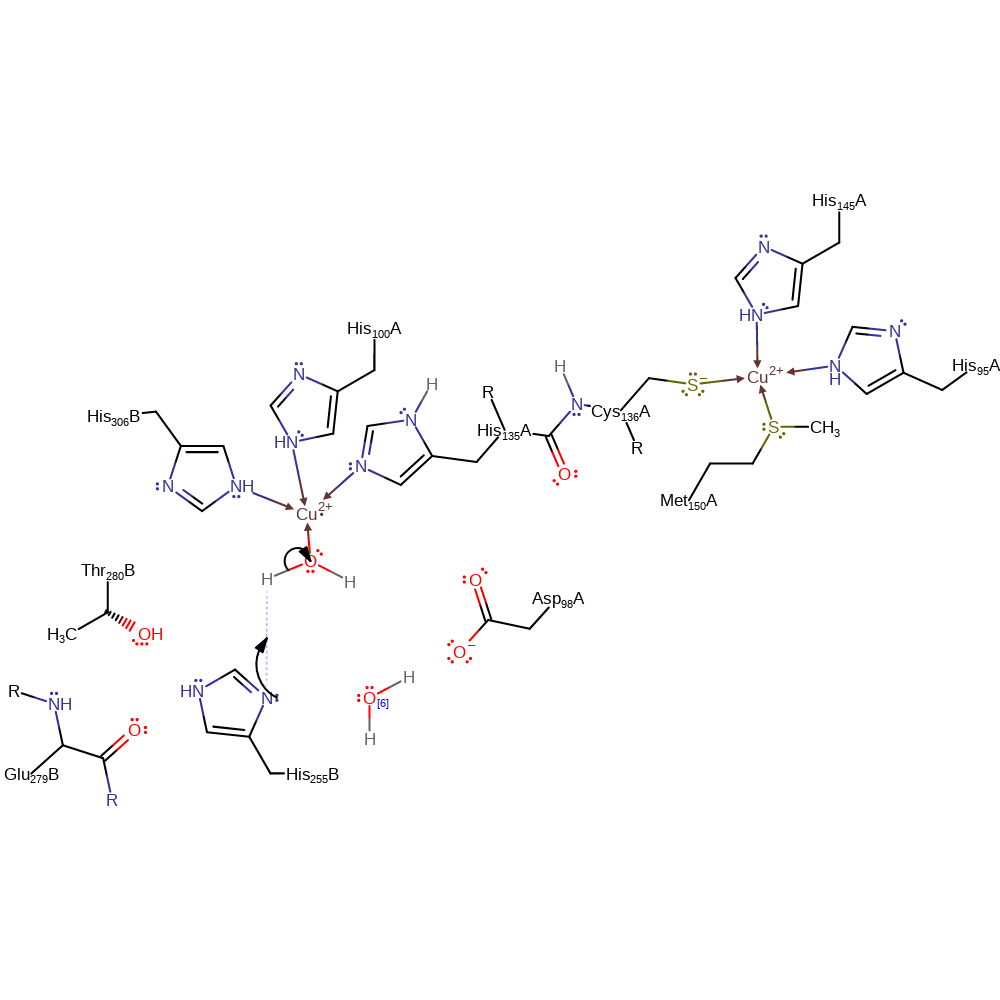
Step 3. Water displaces the NO radical, and is deprotonated by His255B, to regenerate the enzyme's resting state.
Download: Image, Marvin FileCatalytic Residues Roles
| Residue | Roles |
|---|---|
| Asp98A | hydrogen bond acceptor |
| His135A | metal ligand |
| Cys136A | metal ligand |
| His255B | hydrogen bond acceptor |
| Glu279B (main-C) | modifies pKa |
| Thr280B | modifies pKa |
| His255B | proton acceptor |






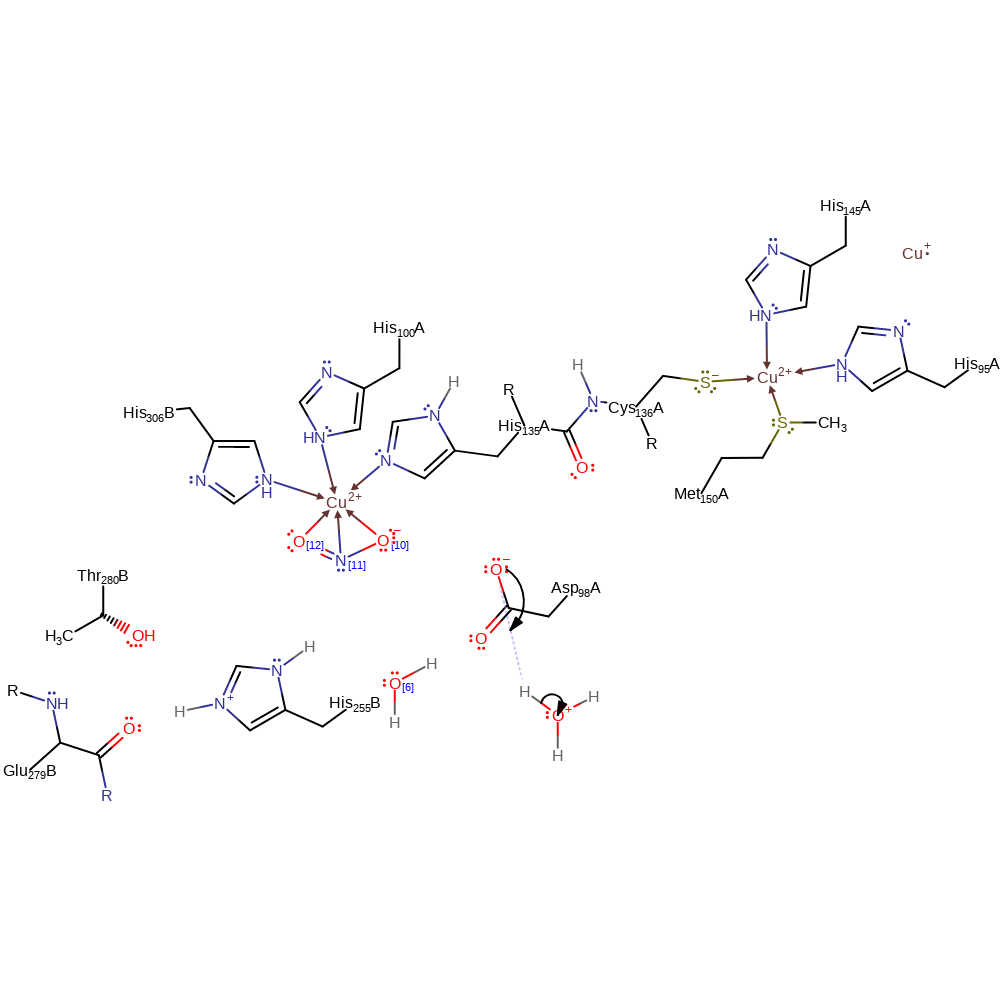



 Download:
Download:  Download:
Download: